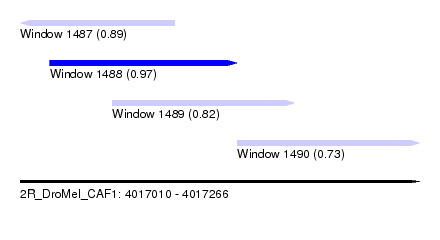
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,017,010 – 4,017,266 |
| Length | 256 |
| Max. P | 0.968939 |

| Location | 4,017,010 – 4,017,109 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.95 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -22.63 |
| Energy contribution | -21.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4017010 99 - 20766785 AAUGCCAAUUUGUUUUAUGGGCGACAAGUUAAGCUUGCCUUUGCUUUGCUUUGAUGUUGAUUUUUAUAUUAUGAUGUCGGAGCGCCCAU--------------GCCCCA-------UCCU ...............((((((((.((((.....)))).)........((((((((((((((......)))..)))))))))))))))))--------------).....-------.... ( -25.20) >DroSim_CAF1 1924 109 - 1 AAUGCCAAUUUGUUUUAUGGGCGACAAGUUAAGCUUGCCUUUGCUUUGCUUUGAUGUUGAUUUUUAUAUUAUGAUGUUGGAGCGCCCAUCCCC--------AUGCCCCCUGUU---UCCC ................(((((((.((((.....)))).)........((((..((((((((......)))..)))))..))))))))))....--------............---.... ( -22.40) >DroYak_CAF1 1706 120 - 1 AAUGCCAAUUUGUUUUAUGGGUGACAAGUUGAGCUUGCCUUUGCUUUGCUUUGAUGUUGAUUUUUAUAUUAUGAUGUCGGAGCGCCCAUCCCCUAUCCCCCAUCCCCCAUCUCCCAUCCU ................(((((.((.((((.((((........)))).))))((((((........)))))).((((..(((..............)))..)))).....)).)))))... ( -21.84) >consensus AAUGCCAAUUUGUUUUAUGGGCGACAAGUUAAGCUUGCCUUUGCUUUGCUUUGAUGUUGAUUUUUAUAUUAUGAUGUCGGAGCGCCCAUCCCC________AUGCCCCAU_U____UCCU ................((((((........((((........)))).((((((((((((((......)))..)))))))))))))))))............................... (-22.63 = -21.97 + -0.66)

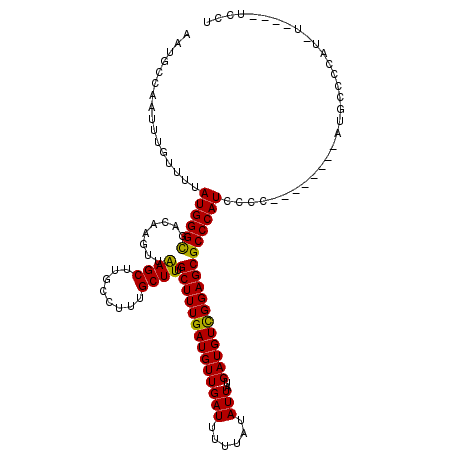
| Location | 4,017,029 – 4,017,149 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4017029 120 + 20766785 CCGACAUCAUAAUAUAAAAAUCAACAUCAAAGCAAAGCAAAGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAA ..(((((((((((((((((((....................((((((.....))))))(((.((.......)).)))))))))).))))))(((.....)))..)))))).......... ( -26.10) >DroSim_CAF1 1953 120 + 1 CCAACAUCAUAAUAUAAAAAUCAACAUCAAAGCAAAGCAAAGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAA ...((((((((((((((((((....................((((((.....))))))(((.((.......)).)))))))))).))))))(((.....)))..)))))........... ( -21.60) >DroYak_CAF1 1746 120 + 1 CCGACAUCAUAAUAUAAAAAUCAACAUCAAAGCAAAGCAAAGGCAAGCUCAACUUGUCACCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAA ..(((((((((((((((((((....................((((((.....))))))..(((..........))).))))))).))))))(((.....)))..)))))).......... ( -23.50) >consensus CCGACAUCAUAAUAUAAAAAUCAACAUCAAAGCAAAGCAAAGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAA ..(((((((((((((((((((....................((((((.....))))))(((.((.......)).)))))))))).))))))(((.....)))..)))))).......... (-21.83 = -22.50 + 0.67)

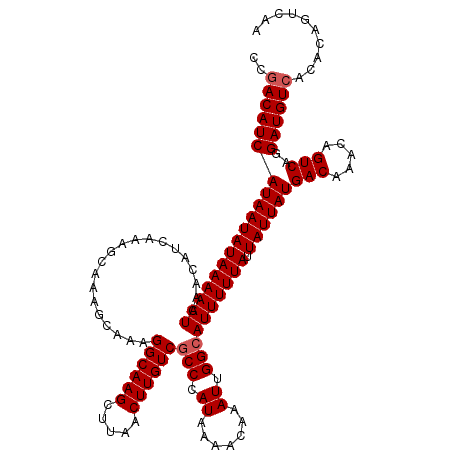
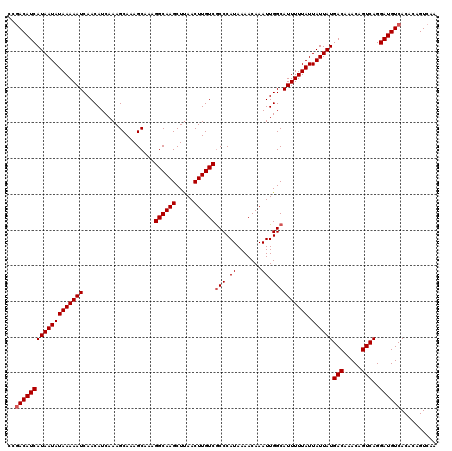
| Location | 4,017,069 – 4,017,186 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 98.86 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4017069 117 + 20766785 AGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAACCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCG .((((((.....))))))................(((.............((((.....))))(((((..............)))))....((((..((....))))))....))). ( -27.94) >DroSim_CAF1 1993 117 + 1 AGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAACCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCG .((((((.....))))))................(((.............((((.....))))(((((..............)))))....((((..((....))))))....))). ( -27.94) >DroYak_CAF1 1786 117 + 1 AGGCAAGCUCAACUUGUCACCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAACCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCG .((((((.....))))))................(((.............((((.....))))(((((..............)))))....((((..((....))))))....))). ( -27.94) >consensus AGGCAAGCUUAACUUGUCGCCCAUAAAACAAAUUGGCAUUUUUAUUAUUAUGACAAACAGUCAGGAUGUCACACAGUCAACCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCG .((((((.....))))))................(((.............((((.....))))(((((..............)))))....((((..((....))))))....))). (-27.94 = -27.94 + -0.00)

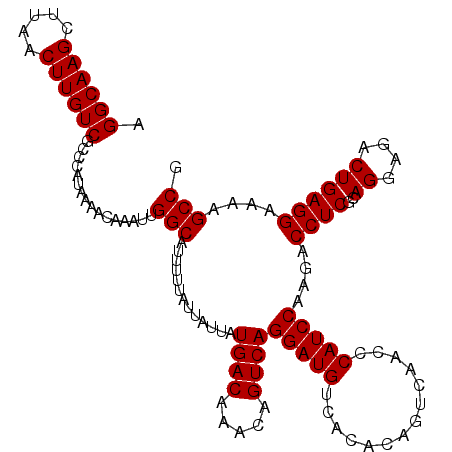
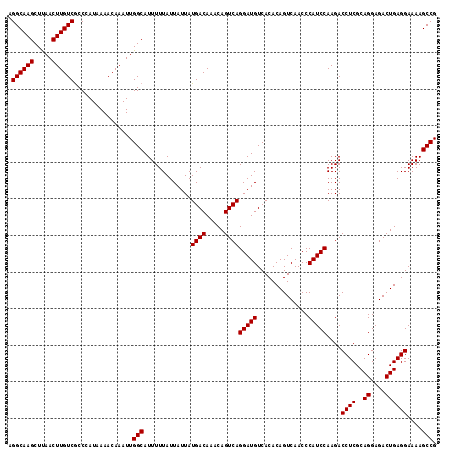
| Location | 4,017,149 – 4,017,266 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -31.56 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4017149 117 + 20766785 CCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCGGAAAAGCGGCGACGGGCCACAAUGAGCUACGCUUAUCUUUUGAAAGCAGCUAAACCGCUUUUGAUUAUGUGGAAGUUGUU (((.((.....((((..((....))))))....((((......)))))).)))(((((((((((...))))))...(..(((((.........)))))..)...)))))........ ( -35.50) >DroSec_CAF1 21 117 + 1 CCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCGGAAAAGCGGCGACGGGCCACAAUGAGCUACGCUUAUCUUUUGAAAGCAGCUAAACUGCUUUUGAUUAUGUGGAAGUUGUU (((.((.....((((..((....))))))....((((......)))))).)))(((((((((((...))))))...(..(((((((.....)))))))..)...)))))........ ( -40.10) >DroSim_CAF1 2073 117 + 1 CCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCGGAAAAGCGGCGACGGGCCACAAUGAGCUACGCUUAUCUUUUGAAAGCAGCUAAACCGCUUUUGAUUAUGUGGAAGUUGUU (((.((.....((((..((....))))))....((((......)))))).)))(((((((((((...))))))...(..(((((.........)))))..)...)))))........ ( -35.50) >DroEre_CAF1 19 116 + 1 CCCAUCCAAGAGCUCGU-GGAGAUUAAGGAAAACCCGGAAAAGCGGCGACGGGCCACAAUGAGCCACGCUUAUCUUUUGAAAGCAGCUAAAGCGCUUUUGAUUAUGUGGAAGUUGUU .((.((((.(....).)-)))......((.....))))......((((((...(((((((((((...))))))...(..(((((.((....)))))))..)...)))))..)))))) ( -32.00) >DroYak_CAF1 1866 117 + 1 CCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCGGAAAAGCGGCGACGGGCCGCAAUGAGCCACGCUUAUCUUUUGAAAGCAGCUAAAGCGCUUUUGAUUAUGUGGAAGUUGUU (((.((.....((((..((....))))))....((((......)))))).)))(((((((((((...))))))...(..(((((.((....)))))))..)...)))))........ ( -38.10) >consensus CCCAUCCAAGACCUCGCAGGAGACUGAGGAAAAGCCGGAAAAGCGGCGACGGGCCACAAUGAGCUACGCUUAUCUUUUGAAAGCAGCUAAACCGCUUUUGAUUAUGUGGAAGUUGUU (((.(((..(....).(((....))).)))...((((......))))...)))(((((((((((...))))))...(..(((((.........)))))..)...)))))........ (-31.56 = -31.32 + -0.24)

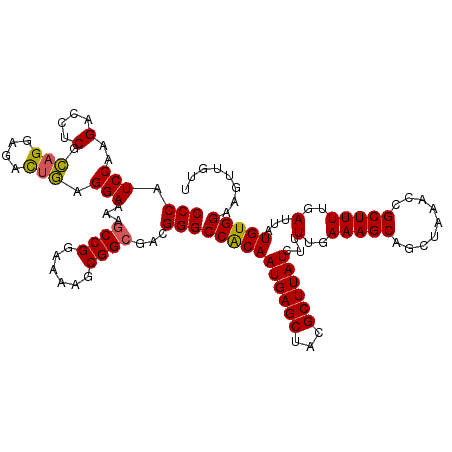
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:09 2006