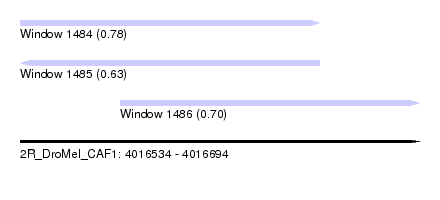
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 4,016,534 – 4,016,694 |
| Length | 160 |
| Max. P | 0.777776 |

| Location | 4,016,534 – 4,016,654 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -30.41 |
| Energy contribution | -30.20 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4016534 120 + 20766785 UCAGCCAUUUUCCAGCUUGUUUUUAAAGUGUCGUAAAUCACGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCUUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAU ......(((((((((............(((........)))(((((((.(..((((((((((((((...(((.....)))...))).))).))))))))..).))))))).))))))))) ( -34.70) >DroSim_CAF1 1439 120 + 1 UCGGCCAUUUUCCAGCUUGUUUUUAAAGUGUCGUAAAUCAUGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAU ......((((((((((((.......))))((.((((((((((..((((........))))......((((((.....(((....)))...)))))).)))))))))).))..)))))))) ( -35.50) >DroYak_CAF1 1204 120 + 1 CCGGCCAUUUUCCAGCAUGUUUUUUAAGUGUCGUAAAUCACGUAGUAGUUUGCAGUUUGUACAGUUCCUACCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAU ......(((((((((............(((........)))(((((((.((..(((((((((((..((..((.....))....)).)))).)))))))..)).))))))).))))))))) ( -27.20) >consensus UCGGCCAUUUUCCAGCUUGUUUUUAAAGUGUCGUAAAUCACGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAU ......(((((((((............(((........)))(((((((.(((((((((((((((..((.(((.....)))...)).)))).))))))))))).))))))).))))))))) (-30.41 = -30.20 + -0.21)

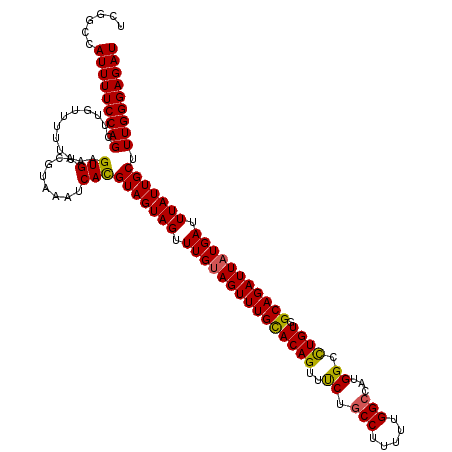
| Location | 4,016,534 – 4,016,654 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -21.17 |
| Energy contribution | -20.40 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4016534 120 - 20766785 AUCUCCCAAAGCAAUAAAUCAUAAUCUGCCACAAGCCAUGGCCAAAAAGGCAGAAACUGUGCAAACUACAAACUACUACGUGAUUUACGACACUUUAAAAACAAGCUGGAAAAUGGCUGA .......((((...((((((((..((((((....((....))......))))))...((((.....)))).........)))))))).....)))).......(((((.....))))).. ( -20.90) >DroSim_CAF1 1439 120 - 1 AUCUCCCAAAGCAAUAAAUCAUAAUCUGCCACAGGCCAUGGCCAAAAAGGCAGAAACUGUGCAAACUACAAACUACUACAUGAUUUACGACACUUUAAAAACAAGCUGGAAAAUGGCCGA .((..(((......((((((((..((((((...(((....))).....))))))...((((.....)))).........))))))))...(((((.......))).)).....)))..)) ( -22.80) >DroYak_CAF1 1204 120 - 1 AUCUCCCAAAGCAAUAAAUCAUAAUCUGCCACAGGCCAUGGCCAAAAAGGUAGGAACUGUACAAACUGCAAACUACUACGUGAUUUACGACACUUAAAAAACAUGCUGGAAAAUGGCCGG .....((...(((.............)))....((((((..(((....(((((....((((.....))))..)))))..(((........))).............)))...)))))))) ( -23.82) >consensus AUCUCCCAAAGCAAUAAAUCAUAAUCUGCCACAGGCCAUGGCCAAAAAGGCAGAAACUGUGCAAACUACAAACUACUACGUGAUUUACGACACUUUAAAAACAAGCUGGAAAAUGGCCGA ..............((((((((..((((((...(((....))).....))))))...((((.....)))).........)))))))).................((((.....))))... (-21.17 = -20.40 + -0.77)

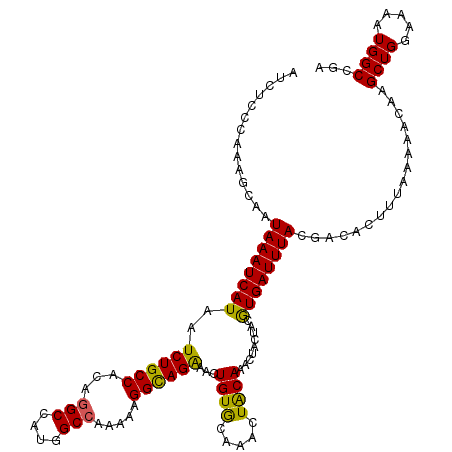
| Location | 4,016,574 – 4,016,694 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -31.47 |
| Energy contribution | -31.37 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 4016574 120 + 20766785 CGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCUUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAUUGCAGAUCCAACUUGCAGGAUGCAGUUAAUUCCUUCGGUA .(((((((.(..((((((((((((((...(((.....)))...))).))).))))))))..).)))))))...(((((((((((..(((........))))))))))..))))....... ( -37.60) >DroSim_CAF1 1479 120 + 1 UGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAUUGCAGCUCCAACUUGCAGGAUGCAGUUAAUUCCUUCGGUA .(((((((.(..((((((((((((((...(((.....)))...)).)))).))))))))..).)))))))...(((((((((((..(((........))))))))))..))))....... ( -37.30) >DroYak_CAF1 1244 120 + 1 CGUAGUAGUUUGCAGUUUGUACAGUUCCUACCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAUUGCAGCUUCAACUUGCCGGUUGCAGUAAAUUUCUUUCGAA .(((((((.((..(((((((((((..((..((.....))....)).)))).)))))))..)).))))))).(((..(((.....(((.(((((....))))).))).....)))..))). ( -31.20) >consensus CGUAGUAGUUUGUAGUUUGCACAGUUUCUGCCUUUUUGGCCAUGGCCUGUGGCAGAUUAUGAUUUAUUGCUUUGGGAGAUUGCAGCUCCAACUUGCAGGAUGCAGUUAAUUCCUUCGGUA .(((((((.(((((((((((((((..((.(((.....)))...)).)))).))))))))))).)))))))...((((.((((((..(((........)))))))))...))))....... (-31.47 = -31.37 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:49:05 2006