| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,991,783 – 3,991,894 |
| Length | 111 |
| Max. P | 0.539094 |

| Location | 3,991,783 – 3,991,894 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -32.15 |
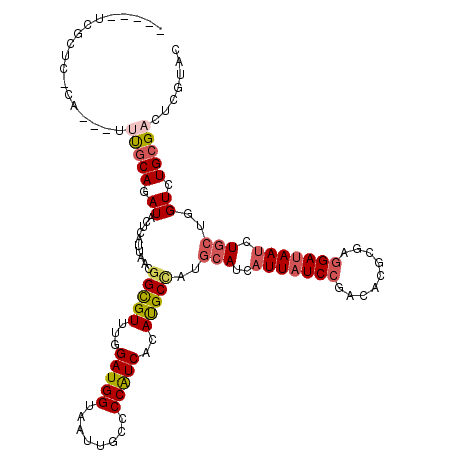
| Consensus MFE | -20.12 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
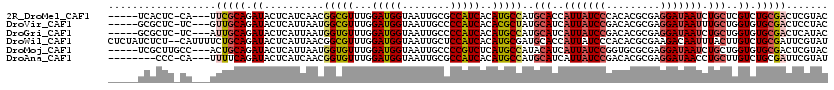
>2R_DroMel_CAF1 3991783 111 - 20766785 -----UCACUC-CA---UUCGCAGAUACUCAUCAACGGCGUUUGGAUGGUAAUUGCGCCAUCACAUGCCAUGCACCAUUAUCCCACACGCGAGGAUAAUCUGCUCGUCUGCGACUCGUAC -----......-..---.((((((((..........(((((...((((((......))))))..)))))..(((..((((((((......).))))))).)))..))))))))....... ( -36.60) >DroVir_CAF1 121 111 - 1 -----GCGCUC-UC---GUUGCAGAUACUCAUUAAUGGCGUUUGGAUGGUAAUUGCCCCAUCACACGCUAUGCAUCAUUAUCCGACACGCGAGGAUAAUUUGCUGGUGUGCGACUCCUAC -----......-..---((((((.((........(((((((...(((((........)))))..)))))))(((..(((((((.........))))))).)))..)).))))))...... ( -34.20) >DroGri_CAF1 121 111 - 1 -----GCGCUC-UC---AUUGCAGAUACUCAUUAAUGGUGUUUGGAUGGUAAUUGCCCCAUCACAUGCCAUGCAUCAUUAUCCGACACGCGAGGAUAAUCUGCUGGUGUGCGACUCAUAC -----((((.(-(.---...((((((........(((((((...(((((........)))))..))))))).......(((((.........))))))))))).)).))))......... ( -32.50) >DroWil_CAF1 121 118 - 1 CUCUAUCUCU--CAUUUUCUGCAGAUACUCAUUAACGGCGUUUGGAUGGUAAUUGCUCCAUCACAUGCGAUGCACCAUUAUCCCACACGCGAAGACAAUUUACUUGUCUGCGAUUCGUAU ...((((((.--........).))))).......(((((((.((((((((.(((((..........)))))..)))))....))).))))(.((((((.....)))))).)....))).. ( -27.00) >DroMoj_CAF1 121 112 - 1 -----UCGCUUGCC---ACUGCAGAUACUCAUUAAUGGUGUUUGGAUGGUAAUUGCCCCGUCUCAUGCCAUACAUCAUUAUCCGGUGCGCGAGGAUAAUCUGCUGGUGUGCGACUCGUAC -----((((.((((---(..((((((.(((....(((((((..((((((........)))))).))))))).((((.......))))...)))....))))))))))).))))....... ( -35.90) >DroAna_CAF1 121 108 - 1 --------CCC-CA---UUUUCAGAUACUCAUCAACGGUGUUUGGAUGGUAAUUGCGCCAUCACAUGCCAUGCAUCAUUAUCCGACACGCGAGGAUAACCUGCUUGUCUGCGAUUCGUAU --------...-..---.......((((..(((...(((((...((((((......))))))..)))))..(((.........((((.((.(((....))))).))))))))))..)))) ( -26.70) >consensus _____UCGCUC_CA___UUUGCAGAUACUCAUUAACGGCGUUUGGAUGGUAAUUGCCCCAUCACAUGCCAUGCAUCAUUAUCCGACACGCGAGGAUAAUCUGCUGGUCUGCGACUCGUAC ..................(((((.((..........(((((...(((((........)))))..)))))..(((..(((((((.........))))))).)))..)).)))))....... (-20.12 = -20.85 + 0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:56 2006