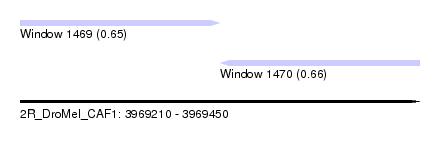
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,969,210 – 3,969,450 |
| Length | 240 |
| Max. P | 0.664756 |

| Location | 3,969,210 – 3,969,330 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3969210 120 + 20766785 CCGAUCGUAUGAACCAGUCGCACUCGUCACUGGUGUGGUUGGGCACAAAGUCCAGAAUGAUUUUGAUGUCCAGCUCUUUGGCGCGCGCAAGCAGGGCCUCGAAAUCCUCCAUUGUGCCGA ............((((((.((....)).))))))..((((((((((((((((......))))))).)))))))))..(((((((((....))((((........)))).....))))))) ( -44.10) >DroVir_CAF1 21465 120 + 1 CAGAGCGUAUAAACCAGUCGCACUCAUCAGACGAGUGGUUGGGCACAAAGUCCAAAAUGAUUUUGAUGUCCAGCUGCUUGGCGCGCUCUAACAACUUUUCAAAGUCCUCCAUGUUGCCAA .(((((((........(((..........)))(((..(((((((((((((((......))))))).))))))))..)))...)))))))..((((.................)))).... ( -39.53) >DroGri_CAF1 16069 120 + 1 CCGAUCGGAUGAACCAAUCGCACUCAUCACUCGAGUGAUUUGGCACAAAGUCCAAAAUAAUUUUAAUAUUCAUCUGGUUGGCUUUCUGCACAAGUGCCUCGAAAUCAGACAUGGUGCCAA (((((((((((((((((...(((((.......)))))..))))...(((((........)))))....)))))))))))))......((((..(((.((.(....))).))).))))... ( -32.70) >DroWil_CAF1 17808 120 + 1 CCGAACGGAUAAACCAAUCACACUCAUCACUAGUGUGAUUAGGCACAAAGUCCAAAAUGAUUUUGAUAUCCAAUUGCUUGGCACGCGCCACCAAAGCAUCAAAAUCUUCCAUGGUGCCAA ......(((((..(((((((((((.......))))))))).))..(((((((......))))))).)))))..(((..((((....)))).))).((((((..........))))))... ( -33.70) >DroMoj_CAF1 22325 120 + 1 CCGAGCGAAUGAACCAAUCGCACUCAUCGGACGAAUGAUUUGGCACAAAGUCCAGGAUGAUUUUGAUGUUCAGCUGAUUGGCGCGCGCCAGCAAUUUAUCAAAGUCCUCCAUGGUGCCAA ((((((((.((...)).)))).....)))).........((((((......((((((.((((((((((....(((....(((....))))))....)))))))))).))).))))))))) ( -37.50) >DroAna_CAF1 15144 120 + 1 CGGAUCUGAUAAACCAGUCGCACUCAUCGCUAGUGUGGUUGGGCACAAAAUCAAGGAUAAUCUUGAUGUCCAGUUCCUUGGCGCGGGCCACUAGUUUCUCAAAGUCCUCCAUGGUGCCGA .(((.(((.....(((..((((((.......))))))..))).......((((((......))))))...))).)))((((((((((..(((..........)))..)))...))))))) ( -36.10) >consensus CCGAUCGGAUAAACCAAUCGCACUCAUCACUCGAGUGAUUGGGCACAAAGUCCAAAAUGAUUUUGAUGUCCAGCUGCUUGGCGCGCGCCACCAAUGCCUCAAAAUCCUCCAUGGUGCCAA ......(((((........((.((.(((((....))))).)))).(((((((......))))))).)))))......(((((((.............................))))))) (-15.74 = -16.13 + 0.39)

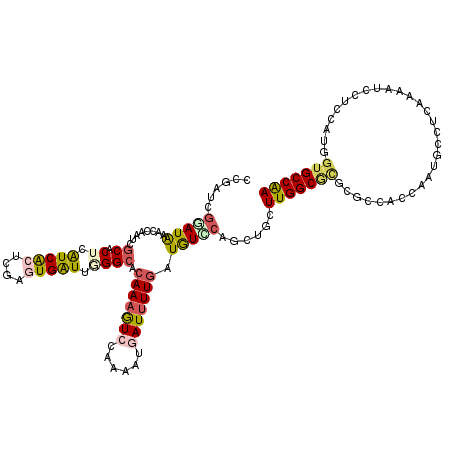
| Location | 3,969,330 – 3,969,450 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -21.34 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
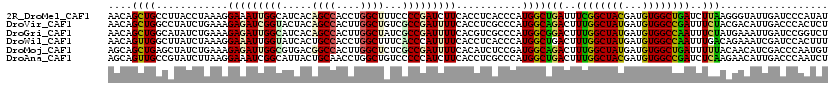
>2R_DroMel_CAF1 3969330 120 - 20766785 AACAGCUGCCUUACCUAAAGGAAAUUGGCAUCACAGCCACCUGGCUUUCCCCGAUCUUCACCUCACCCAUGGCUGAUUUCGGCUACGAUGUGGCUGAUCUUAAGGGUAUUGAUCCCAUAU ..(((((.((((.....))))......(((((..((((....))))....((((...(((((........)).)))..))))....)))))))))).......(((.......))).... ( -33.80) >DroVir_CAF1 21585 120 - 1 AACAGCUGGCCUAUCUGAAAGAGAUCGGUACUACAGCCACUUGGCUGUCGCCGAUUUUCACCUCGCCCAUGGCAGACUUUGGCUAUGAUGUGGCCGAUUUCUACGACAUUGACCCACUCU ....((((((.....((((...(((((((...((((((....)))))).)))))))))))....)))...)))(((..((((((((...))))))))..))).................. ( -37.50) >DroGri_CAF1 16189 120 - 1 AACAGCUGGCAUAUCUGAAAGAGAUUGGCAUCACAGCCACUUGGCUAUCGCCGAUUUUCACGUCGCCCAUGGCGGACUUUGGCUAUGAUGUGGCCAAUUUCUAUGAAAUUGAUCCGGUCU ....(((((((..((....((((((((((((((.(((((.(((((....))))).......((((((...))).)))..))))).))))...))))))))))..))...))..))))).. ( -39.90) >DroWil_CAF1 17928 120 - 1 AACAGUUGGCUUAUCUAAAGGAAAUUGGUAUCACUGCCACCUGGCUUUCACCCAUUUUCACCUCACCCAUGGCUGACUUUGGCUAUGAUGUGGCCAAUUUGACAGAAAUCGAUCCACUUU ...(((((((.........((((((.(((......(((....)))....))).))))))....(((.(((((((......)))))))..))))))))))..................... ( -30.20) >DroMoj_CAF1 22445 120 - 1 AGCAGCUGAGCUAUCUGAAAGAGAUUGGCGUGACGGCCACUUGGCUCUCGCCGAUUUUCACAUCUCCGAUGGCAGACUUUGGCUAUGAUGUGGCUGAUUUUUACAACAUCGACCCAAUGU .........((((((.((..((((((((((.((..(((....))))).))))))))))......)).))))))..((.((((...((((((..............))))))..)))).)) ( -34.64) >DroAna_CAF1 15264 120 - 1 AGCAGUUGCCGUAUCUUAAGGAAAUCGGCAUUACUGCAACCUGGCUGUCCCCCAUCUUCACCUCGCCCAUGGCUGACUUUGGCUACGAUGUGGCCGAUCUCAAGAACAUUGACCCAAUCU .((((((((((..((.....))...)))))..)))))....(((..(((.....((((......(((...))).((..((((((((...))))))))..)))))).....)))))).... ( -34.70) >consensus AACAGCUGGCCUAUCUGAAAGAAAUUGGCAUCACAGCCACCUGGCUGUCGCCGAUUUUCACCUCGCCCAUGGCUGACUUUGGCUAUGAUGUGGCCGAUUUCAACGACAUUGACCCAAUCU ....((((............(((((((((.....((((....))))...)))))))))...........))))(((..((((((((...))))))))..))).................. (-21.34 = -20.82 + -0.52)
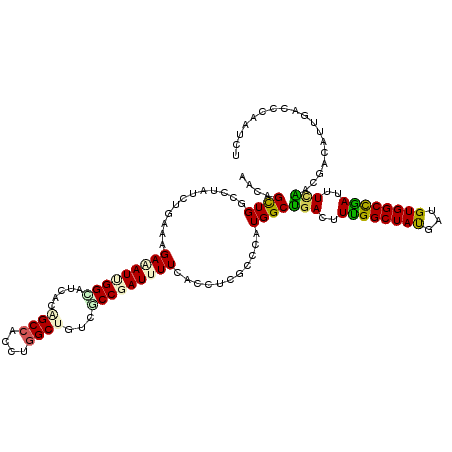
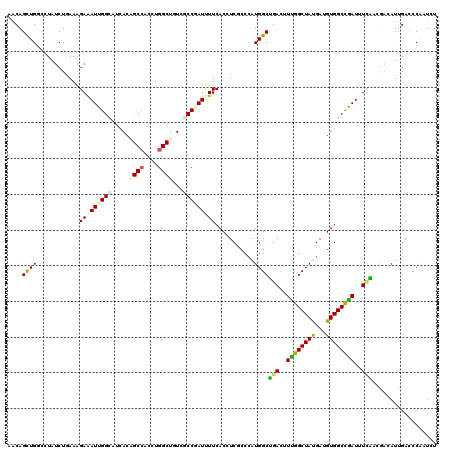
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:50 2006