| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,940,047 – 3,940,152 |
| Length | 105 |
| Max. P | 0.621938 |

| Location | 3,940,047 – 3,940,152 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
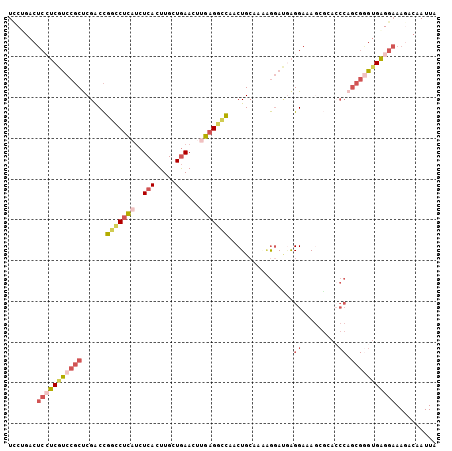
| Mean pairwise identity | 78.73 |
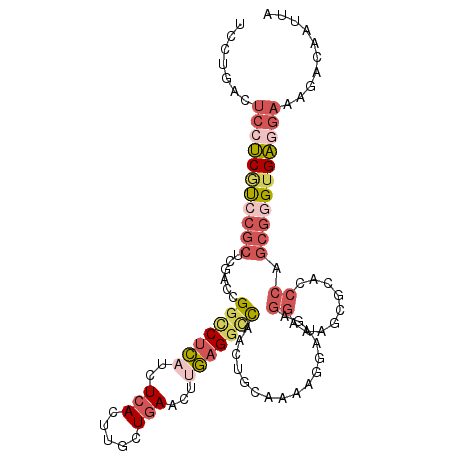
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -19.04 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3940047 105 - 20766785 UUCGGACUCCUCGUCCGCUCGACCGUCCUCAUCUCACUUGCUAAACUUGAGGCCCACUGCAAAAGGAUAAGGAAAGCGCACCCAGCGAGAGAGGAAAGACAAUUA ..(((((.....))))).....(..(((((.((((....(((...((((...((..........)).))))...)))((.....)))))))))))..)....... ( -26.70) >DroSec_CAF1 653 105 - 1 UCCUGACUCGUCGUCCGCUCGACCGGCCUCAUCUCACUUGCUGAACUUGAGGCCCACUGCAAAAGGACGAGGAAAACGCACCCAGCGGGUGAGGAAAUACAAUUA ((((.((((.((((((((......(((((((..(((.....)))...)))))))....))....)))))).))...(((.....))).)).)))).......... ( -37.00) >DroSim_CAF1 1746 105 - 1 UCCGGACUCCUCGUCCGCUCGACCGGCCUCAUCUCACUUGCUGAACUUGAGGCCCACUGCAAAAGGACGAGGAAAGCGCACCCAGCGGGUGAGGAAAGACAAUUA (((....(((((((((((......(((((((..(((.....)))...)))))))....))....))))))))).....(((((...))))).))).......... ( -40.40) >DroEre_CAF1 592 105 - 1 UCCUGACUCCUCGUCCGCUCGACCGGCCUCCUCUCACUUGCUGAACUUGUGGUUACCUGGAACAGGAUGAGGAAAGGAAAACCAGCGGGUGAGGAAAGACAAUUA ..((...(((((..(((((.......(((((((((((..(.....)..))))...((((...))))..)))))..))......)))))..))))).))....... ( -33.52) >DroYak_CAF1 1632 104 - 1 UCCUGACUCCUCGUCCGCCCGACCGGUCUCAUCUCACUUGCUGAACUUGAGGCCAACUGCAAC-GGAUUAGGAUAGAGGACCCAGCGGGUGAGGAAAGACAAUUA ..((...(((((..((((....(((((((((..(((.....)))...)))))))..(((...(-......)..))).)).....))))..))))).))....... ( -31.50) >DroPer_CAF1 554 93 - 1 UCCACAC-CAACACCCCCUCGGCUGGCCUGCACUCACUUGCUGAACUUCAGACCAACUGCAAGGAAAGGGGGUAA----A-------AGUGUGACAAAAUAAUUA ..(((((-....(((((((((((.((..........)).))))).((((((.....))).)))....))))))..----.-------.)))))............ ( -27.60) >consensus UCCUGACUCCUCGUCCGCUCGACCGGCCUCAUCUCACUUGCUGAACUUGAGGCCAACUGCAAAAGGAUGAGGAAAGCGCACCCAGCGGGUGAGGAAAGACAAUUA .......(((((((((((......(((((((..(((.....)))...)))))))................((.........)).))))))))))).......... (-19.04 = -20.88 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:41 2006