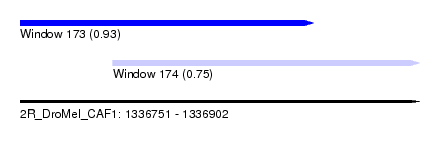
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,336,751 – 1,336,902 |
| Length | 151 |
| Max. P | 0.931091 |

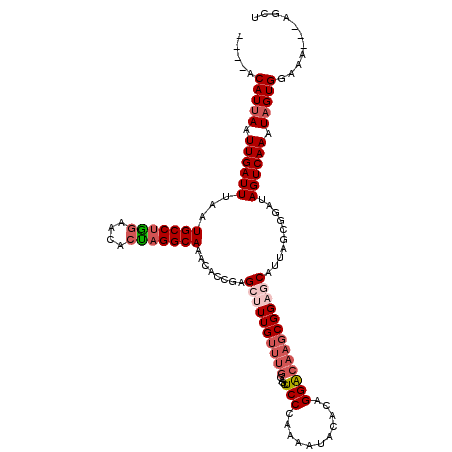
| Location | 1,336,751 – 1,336,862 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.96 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1336751 111 + 20766785 AGUUCU-GUGUUUUACACCCCGGCGGACAAACUCAC--------ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACAUCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAA .(((((-((((.........(..(((((((((((..--------.((.....))......(((((((....)))))))......))).))))))))..)..........))))))))).. ( -32.81) >DroSec_CAF1 55271 112 + 1 AGUUCUGGUGUUUUACACCCCGGCGGACAAACUCAC--------ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACAGCGAGCCUUGUUUGGAGCUCCCAAAAUACACAGGGCAA .(((((((((((((......(..(((((((.(((.(--------.((.....))......(((((((....)))))))....).)))..)))))))..).....))))))).)))))).. ( -31.00) >DroSim_CAF1 33470 112 + 1 AGUUCUGGUGUUUUACACCCCGGCGGACAAACUCAC--------ACAUUAAUUGAUUUAAUGCCCGGAACACUAGGCAAACAGCGAGCCUUGUUUGGAGCUCCCAAAAUACACAGGGCAA ....((((((((.......((((.(((.....))..--------.((((((.....)))))))))))))))))))((.....))..(((((((((((.....)))).....))))))).. ( -29.51) >DroEre_CAF1 36680 120 + 1 AGUUCUGUCGCUCUACCUCCCGGCGGACGAACUCACACUUACACCCAUUAAUUGAUUUAAUGCCUGGAACAGCAGGCAAAUGCCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAA .(((((((.((((....(((.(((((..................))(((((.....)))))))).)))......(((....)))))))....(((((.....)))))....))))))).. ( -32.67) >DroYak_CAF1 23905 108 + 1 AGUUCUGGUGCUUUAUGUCCAGACGGAUAAGCUCAC--------GCAUUAAUUGAUUUAAUGCCUAGAACACUAGGCAAAUACCGAGCUUUG----AGUCUCCGAAAAUACACAGGACAA .(((((((((.(((......((((....((((((..--------..(((((.....)))))((((((....)))))).......))))))..----.))))...))).))).)))))).. ( -32.70) >consensus AGUUCUGGUGUUUUACACCCCGGCGGACAAACUCAC________ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACACCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAA .(((((((((((((......(..(((((((((((............(((((.....)))))((((((....)))))).......))).))))))))..).....))))))).)))))).. (-23.44 = -24.96 + 1.52)

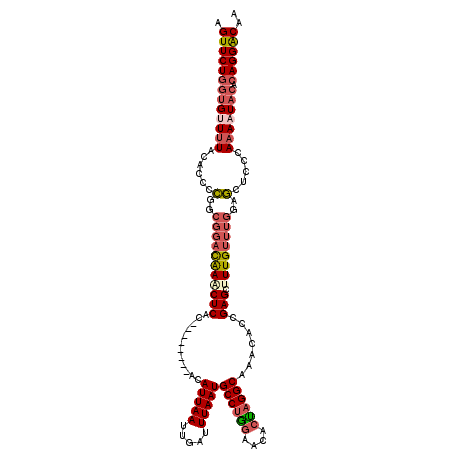
| Location | 1,336,786 – 1,336,902 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -22.80 |
| Energy contribution | -24.44 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1336786 116 + 20766785 ----ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACAUCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAAGCGGAGCAUUAGCGGAUAGUCAAAUAGUGGAAAUCGAGCU ----.(((((.((((((...(((((((....)))))))........((((((((((....(((...........)))))))))))))..........)))))).)))))........... ( -30.90) >DroSec_CAF1 55307 106 + 1 ----ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACAGCGAGCCUUGUUUGGAGCUCCCAAAAUACACAGGGCAAGCGGAGCAUUAGCGGAUAGUCAAAUAGUGG---------- ----.(((((.((((((...(((((((....)))))))....((..(((((((((((.....)))).....)))))))..))...((....))....)))))).))))).---------- ( -34.90) >DroSim_CAF1 33506 106 + 1 ----ACAUUAAUUGAUUUAAUGCCCGGAACACUAGGCAAACAGCGAGCCUUGUUUGGAGCUCCCAAAAUACACAGGGCAAGCGGAGCAUUAGCGGAUAGUCAAAUAGUGG---------- ----.(((((.(((((((((((((((.........((.....))..(((((((((((.....)))).....)))))))...))).))))))......)))))).))))).---------- ( -32.20) >DroEre_CAF1 36720 120 + 1 ACACCCAUUAAUUGAUUUAAUGCCUGGAACAGCAGGCAAAUGCCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAAGCGGAACAUGGACGGAUAGUCAAGUAGUGGAAAUCGAGCU ....((((((.((((((...((((((.....((.(((....)))(((((((....)))))))..................))....)).)).))...)))))).)))))).......... ( -31.70) >DroYak_CAF1 23941 112 + 1 ----GCAUUAAUUGAUUUAAUGCCUAGAACACUAGGCAAAUACCGAGCUUUG----AGUCUCCGAAAAUACACAGGACAAGCGGAACAUUAGCGGAUAGUCAAGUUGUGGAAAGGCAGCU ----((..............(((((((....)))))))....((((((((.(----(.(.((((..(((.....(......).....)))..)))).).))))))).)))....)).... ( -25.20) >consensus ____ACAUUAAUUGAUUUAAUGCCUGGAACACUAGGCAAACACCGAGCUUUGUUUGGAGCUCCCAAAAUACACAGGACAAGCGGAGCAUUAGCGGAUAGUCAAAUAGUGGAAA___AGCU .....(((((.((((((...(((((((....)))))))........((((((((((....(((...........)))))))))))))..........)))))).)))))........... (-22.80 = -24.44 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:20 2006