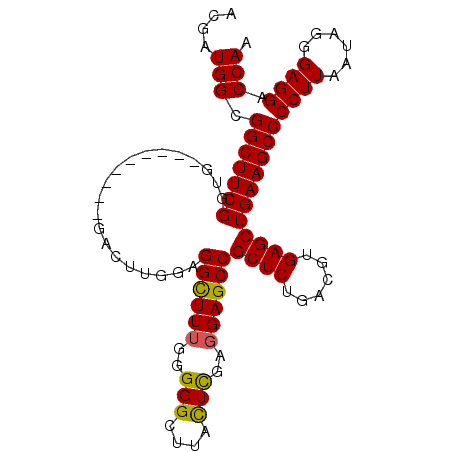
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,870,389 – 3,870,516 |
| Length | 127 |
| Max. P | 0.804752 |

| Location | 3,870,389 – 3,870,498 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -22.59 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3870389 109 + 20766785 AAAUUCCACUUUCAUUAAA---AGUUUCGGCUAAUGUUUUGGUCCUCCCUACUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCGAAUAAACCCCCAAAACCUCCAAGUC------ .......(((((.....))---)))...((((...(((((((............(((((((...((((......)))))))))))...........))))))).....))))------ ( -23.50) >DroSec_CAF1 126146 109 + 1 AAAUUCCAUUUUCAUUAAA---AGUUUUGGCUAAUGUUUUGGUCCUCCCUAUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCGAAUAAGCCCCCAAAGCCUCCAAGUC------ ...................---...(((((.....(((((((.........(..(((((((...((((......)))))))))))..)........)))))))..)))))..------ ( -24.83) >DroEre_CAF1 127156 118 + 1 AAAUUCCGUUUUCAUUCAAAGAAGUUUUGGCUAAUGUUUUGGUCCUCCCUUUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCAAGUAAGCCCCCCAAGCCUCCAAAUCCUCCAC ...................(((((....((((((....))))))....)))))..(((((((..((((......))))((((.........))))....))))).))........... ( -25.90) >DroYak_CAF1 124518 112 + 1 AAAUUCCAUUUUCAUUAAAACAAGUUUUGGCCAAUGUUUUGGUCCUCUUUAUUAAGGGGCUUCAGCUCACGUAAGAGCGGCUCCUCAAGCAAGCCCCCAAAGCCUCCAAAUC------ .......................(((((((((((....)))))............(((((((..((((......)))).(((.....)))))))))))))))).........------ ( -29.60) >consensus AAAUUCCAUUUUCAUUAAA___AGUUUUGGCUAAUGUUUUGGUCCUCCCUAUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCAAAUAAGCCCCCAAAGCCUCCAAAUC______ .........................(((((.....(((((((............(((((((...((((......)))))))))))...........)))))))..)))))........ (-22.59 = -22.65 + 0.06)


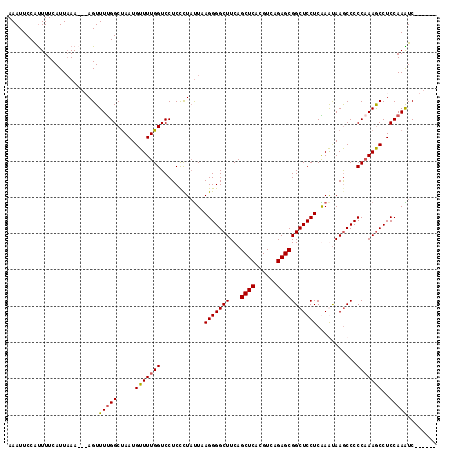
| Location | 3,870,424 – 3,870,516 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3870424 92 + 20766785 UUGGUCCUCCCUACUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCGAAUAAACCCCCAAAACCUCCAAGUC---------CACCGAAGCCACCAUCGU .((((....(((....)))((((((.((((......))))((((..........................))))---------....)))))))))).... ( -22.07) >DroSec_CAF1 126181 92 + 1 UUGGUCCUCCCUAUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCGAAUAAGCCCCCAAAGCCUCCAAGUC---------CACCGAAGCCGCCAUCGU .((((....(((....)))((((((.((((......))))((((.........)))).................---------....)))))))))).... ( -26.20) >DroEre_CAF1 127194 101 + 1 UUGGUCCUCCCUUUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCAAGUAAGCCCCCCAAGCCUCCAAAUCCUCCACAUCCGCCGAAGCCGCCAUCGU .((((....(((....)))((((((.((((......))))((((.........))))..............................)))))))))).... ( -26.00) >DroYak_CAF1 124556 92 + 1 UUGGUCCUCUUUAUUAAGGGGCUUCAGCUCACGUAAGAGCGGCUCCUCAAGCAAGCCCCCAAAGCCUCCAAAUC---------CACCGAAGCCUCCAUCGU .................((((((((.((((......))))((((.........)))).................---------....)))))).))..... ( -23.80) >consensus UUGGUCCUCCCUAUUAAGGGGCUUCAGCUCACGUCAGAGCGGCUCCUCAAAUAAGCCCCCAAAGCCUCCAAAUC_________CACCGAAGCCGCCAUCGU .(((((((........)))((((((.((((......))))((((.........))))..............................)))))))))).... (-23.02 = -23.40 + 0.38)

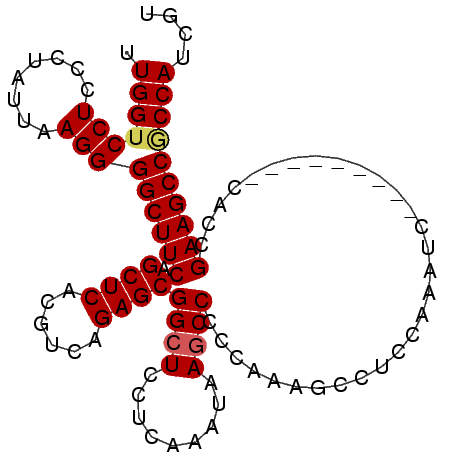
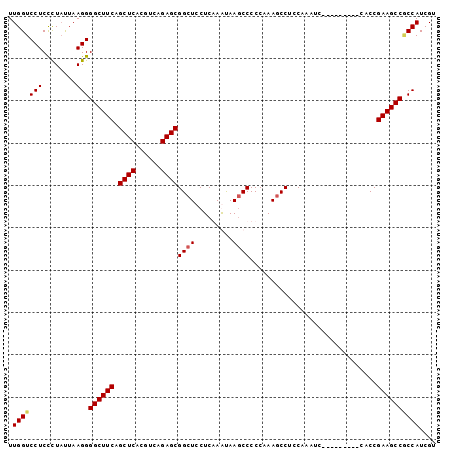
| Location | 3,870,424 – 3,870,516 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -34.65 |
| Consensus MFE | -30.51 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3870424 92 - 20766785 ACGAUGGUGGCUUCGGUG---------GACUUGGAGGUUUUGGGGGUUUAUUCGAGGAGCCGCUCUGACGUGAGCUGAAGCCCCUUAGUAGGGAGGACCAA ....(((((((((((((.---------.(((..((((((((..(((....)))..))))))..))..).))..)))))))))((((......)))))))). ( -32.60) >DroSec_CAF1 126181 92 - 1 ACGAUGGCGGCUUCGGUG---------GACUUGGAGGCUUUGGGGGCUUAUUCGAGGAGCCGCUCUGACGUGAGCUGAAGCCCCUUAAUAGGGAGGACCAA ....(((.(((((((((.---------..((((((((((.....))))..))))))..)))((((......)))).))))))((((......)))).))). ( -35.30) >DroEre_CAF1 127194 101 - 1 ACGAUGGCGGCUUCGGCGGAUGUGGAGGAUUUGGAGGCUUGGGGGGCUUACUUGAGGAGCCGCUCUGACGUGAGCUGAAGCCCCUUAAAAGGGAGGACCAA ....(((.(((((((((..((((((((....(((...((..((.......))..))...))))))).))))..)))))))))((((......)))).))). ( -37.40) >DroYak_CAF1 124556 92 - 1 ACGAUGGAGGCUUCGGUG---------GAUUUGGAGGCUUUGGGGGCUUGCUUGAGGAGCCGCUCUUACGUGAGCUGAAGCCCCUUAAUAAAGAGGACCAA ....(((.(((((((...---------........((((((..(((....)))..))))))((((......)))))))))))((((......)))).))). ( -33.30) >consensus ACGAUGGCGGCUUCGGUG_________GACUUGGAGGCUUUGGGGGCUUACUCGAGGAGCCGCUCUGACGUGAGCUGAAGCCCCUUAAUAGGGAGGACCAA ....(((.(((((((....................((((((..(((....)))..))))))((((......)))))))))))((((......)))).))). (-30.51 = -30.07 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:15 2006