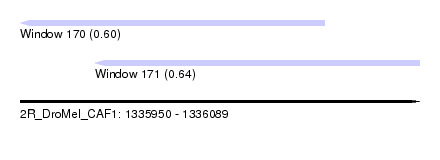
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,335,950 – 1,336,089 |
| Length | 139 |
| Max. P | 0.642878 |

| Location | 1,335,950 – 1,336,056 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.68 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -27.06 |
| Energy contribution | -27.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1335950 106 - 20766785 ACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCGCCUGCCACUCCGCUGGCUCUGG------------CCUGCCA-- ..((((..(.(((.((((.((((..((.((.((((((((......(((((((((.......))))))))))))))))).)).))..))))))))))).)------------..)))).-- ( -40.90) >DroSec_CAF1 54466 106 - 1 ACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCUGCCACUCCGCCGGCUCUGG------------CCUGACA-- ..(((.....(((.((((.((((..((.((.(.((((((......(((((((((.......))))))))))))))).).)).))..))))))))))).)------------)).....-- ( -35.10) >DroSim_CAF1 32655 106 - 1 ACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCUGCCACUCCGCCGGCUCUGG------------CCUGACA-- ..(((.....(((.((((.((((..((.((.(.((((((......(((((((((.......))))))))))))))).).)).))..))))))))))).)------------)).....-- ( -35.10) >DroEre_CAF1 35963 120 - 1 UCGGCAAUUGAGAUGCUGUGUGGUGUGCUAAGUGCUGUCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCUGCCACUCCGCUGGCUCUGACUCCGGCUCUGGCCUGACAGA ((((....(.(((.((((((..(((.((...(.((((((......(((((((((.......))))))))))))))))...)))))..))).)))))).)..)))).((((......)))) ( -35.70) >DroYak_CAF1 23155 106 - 1 ACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUAAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCUGCCACUCCGCUGGAUCUAA------------CCUGAUA-- ((((((.......))))))..(((((((..(((((.(.((((((....((((((.......))))))..)))))).)...).))))..)).)))))...------------.......-- ( -25.50) >consensus ACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCUGCCACUCCGCUGGCUCUGG____________CCUGACA__ ..........(((.((((.((((..((.((.(.((((((......(((((((((.......))))))))))))))).).)).))..)))))))))))....................... (-27.06 = -27.06 + 0.00)


| Location | 1,335,976 – 1,336,089 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -30.80 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
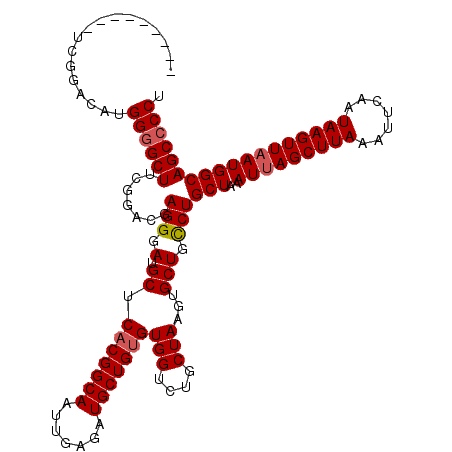
>2R_DroMel_CAF1 1335976 113 - 20766785 -------CAUCGGACAUGGGGCUUCGGACGAGGGAUGCUCACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCGCCU -------...(((((......((((....))))....(.(((((((.......))))))).))))))....((((((((......(((((((((.......))))))))))))))))).. ( -37.60) >DroSec_CAF1 54492 120 - 1 CCUGGGACUUCGGACAUGGGGCUUCGGUCGAGGGAUGCUCACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCU ...((((((((((((....(((....)))........(.(((((((.......))))))).))))))..))))((((((......(((((((((.......)))))))))))))))))). ( -42.00) >DroSim_CAF1 32681 120 - 1 UCUGGGAAUUCGGACAUGGGGCUUCGGUCGAGGGAUGCUCACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCU ...(((....(((((....(((....)))........(.(((((((.......))))))).))))))......((((((......(((((((((.......)))))))))))))))))). ( -40.00) >DroEre_CAF1 36003 111 - 1 ---------UCGGACCCGGGGCUUUGGGCGAGGGAAGCUCUCGGCAAUUGAGAUGCUGUGUGGUGUGCUAAGUGCUGUCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCU ---------........((((((..(((((((((...)))))((((....(((((((....))))).))...))))))))(((..(((((((((.......)))))))))))))))))). ( -36.40) >DroYak_CAF1 23181 111 - 1 ---------UCGGACAUGGGGCUUUGCACAAGGGAUGCUCACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUAAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCU ---------........(((((...((((..((.(.((((((((((.......))))))).))).).))..))))....(((((....((((((.......))))))..)))))))))). ( -36.00) >consensus _________UCGGACAUGGGGCUUCGGACGAGGGAUGCUCACGGCAAUUGAGAUGCUGUGUGGUCUGCUAAGUGCUGCCUGCUAAAUUAGCUUAAAUUCAAUAAGUUAAUGGCAGCCCCU .................((((((.......(((.(.((.(((((((.......)))))))(((....)))...))).)))(((..(((((((((.......)))))))))))))))))). (-30.80 = -31.24 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:16 2006