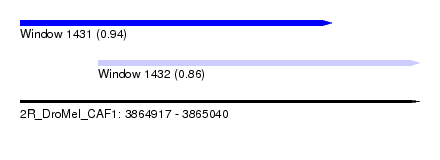
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,864,917 – 3,865,040 |
| Length | 123 |
| Max. P | 0.937468 |

| Location | 3,864,917 – 3,865,013 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -29.73 |
| Energy contribution | -29.60 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
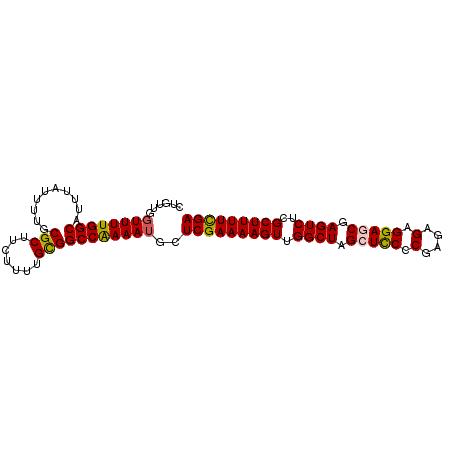
>2R_DroMel_CAF1 3864917 96 + 20766785 CUGUUGGUUUUGGCAUUUAUUUUGCGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUCCCCGAGAGAGGAGCGAGUCUCGCUUUUCGA ......((((((((..........(((.......)))))))))))..(((((((((.((((.(((((.(....).))))).))))..))))))))) ( -36.60) >DroSec_CAF1 120678 96 + 1 CUGUUGGUUUUGGCAUUUAUUUUGCGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUUCCCGAGAGAGGAGCGAGUCGCGCUUUUCGA ......((((((((..........(((.......)))))))))))..((((((((((((((.(((((.(....).))))).))))).))))))))) ( -34.30) >DroEre_CAF1 121839 96 + 1 CUGUUGGUUUUGGCAUUUAUUUUGCGCUUCUUUUGCGGCCGAAAAGCUCGAAAAGUUGGCUAGGUUCCCGAGAGAGGAGCGAGUCUCGCUUUUUGA ..((...(((((((..........(((.......)))))))))).))(((((((((.((((..((((((....).))))).))))..))))))))) ( -29.80) >DroYak_CAF1 118948 96 + 1 CUGUUGGUUUUGGCAUUUAUUUUGCGCUUCUUUUGUGGCCAAAAUGCUCGAAAAGUUGGCUAGGUCCCCGAGAGAGGAACGAGUCUCGCUUUUUGA ......((((((((..........(((.......)))))))))))..(((((((((.((((...(((.(....).)))...))))..))))))))) ( -27.00) >consensus CUGUUGGUUUUGGCAUUUAUUUUGCGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUCCCCGAGAGAGGAGCGAGUCUCGCUUUUCGA ......((((((((..........(((.......)))))))))))..(((((((((.((((.(((((.(....).))))).))))..))))))))) (-29.73 = -29.60 + -0.13)

| Location | 3,864,941 – 3,865,040 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -33.89 |
| Energy contribution | -33.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
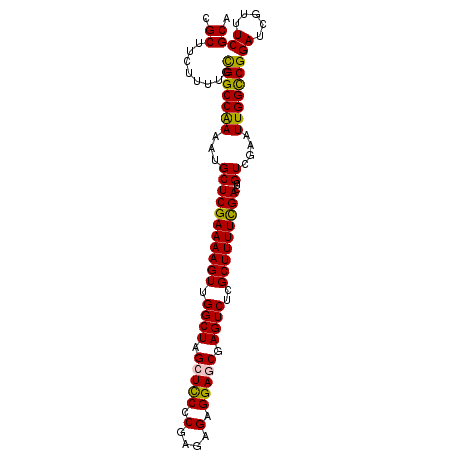
>2R_DroMel_CAF1 3864941 99 + 20766785 CGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUCCCCGAGAGAGGAGCGAGUCUCGCUUUUCGACUGUCGGAUUGGCCGGAUCGUUUCGCA .((........(((((((..((((((((((((.((((.(((((.(....).))))).))))..)))))))))..).))..)))))))((.....)))). ( -41.50) >DroSec_CAF1 120702 99 + 1 CGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUUCCCGAGAGAGGAGCGAGUCGCGCUUUUCGACUGUCGGAUUGGCCGGAUCGUUUCGCA .((........(((((((..(((((((((((((((((.(((((.(....).))))).))))).)))))))))..).))..)))))))((.....)))). ( -39.20) >DroEre_CAF1 121863 99 + 1 CGCUUCUUUUGCGGCCGAAAAGCUCGAAAAGUUGGCUAGGUUCCCGAGAGAGGAGCGAGUCUCGCUUUUUGACUGUCGAAUUGGCCGGAUCGUUUCGCA .((........(((((((..((.(((((((((.((((..((((((....).))))).))))..)))))))))))......)))))))((.....)))). ( -35.70) >DroYak_CAF1 118972 99 + 1 CGCUUCUUUUGUGGCCAAAAUGCUCGAAAAGUUGGCUAGGUCCCCGAGAGAGGAACGAGUCUCGCUUUUUGACUGUCGAAUUGGUCGGAUCGUUUCGCA .((.(((((((.((((.....((.(((....)))))..)).)).)))))))(((((((.(((.(((.((((.....))))..))).)))))))))))). ( -29.60) >consensus CGCUUCUUUUGCGGCCAAAAUGCUCGAAAAGUUGGCUAGCUCCCCGAGAGAGGAGCGAGUCUCGCUUUUCGACUGUCGAAUUGGCCGGAUCGUUUCGCA .((........(((((((...(((((((((((.((((.(((((.(....).))))).))))..)))))))))..))....)))))))((.....)))). (-33.89 = -33.32 + -0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:11 2006