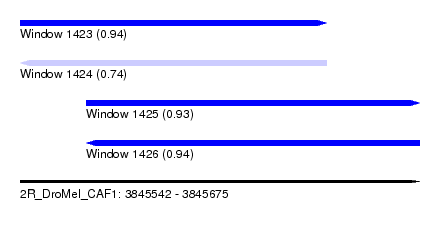
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,845,542 – 3,845,675 |
| Length | 133 |
| Max. P | 0.942550 |

| Location | 3,845,542 – 3,845,644 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -25.66 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
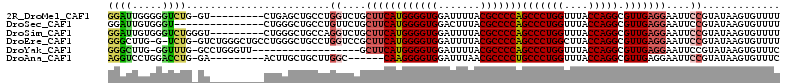
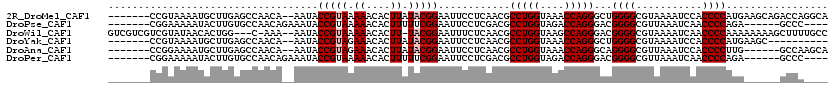
>2R_DroMel_CAF1 3845542 102 + 20766785 GGAUUGGGGGUCUG-GU---------CUGAGCUGCCUGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUU ......(((((...-.(---------((.(((.(((((((..(((...(((((((.......))))))).....)))..)))))))))).))).)))))............. ( -39.40) >DroSec_CAF1 101474 97 + 1 GGAUUGUGGGU---------------CUGGGCUGCCUGUUCUGCUUCAUGGGGUGGACUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUU (((..(..(((---------------.......)))..)....((((((((((((.......)))))))(((((((....))))).)))))))...)))............. ( -32.10) >DroSim_CAF1 118936 103 + 1 GGAUUGUGGGUCUGGGU---------CUGGGCUGCCAGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUU (((..(..(..((((((---------....))..))))..)..)(((((((((((.......)))))))(((((((....))))).))))))....)))............. ( -39.30) >DroEre_CAF1 102767 109 + 1 GGGCUUG-G-UCUG-GUCUGGGCUGCCUGGGCUGCCUGGUCCGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGCUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUU ..(((((-(-(..(-(((.((((((...((((.....(((((((((....)))))))))....)))))))))).)))).))))))).....((....))............. ( -48.30) >DroYak_CAF1 99748 92 + 1 GGGCUUG-GGUUUG-GCCUGGGUU------------------GCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUC (((((..-.....)-))))(((..------------------.((((((((((((.......)))))))(((((((....))))).)))))))...)))............. ( -36.50) >DroAna_CAF1 97270 96 + 1 AGGUCCUGGACCUG-GA---------ACUUGCUGCUUGGC------CAAGGGGUGGAUUUAACGCCCCUGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUC (((((...))))).-..---------(((((.((((..((------..(((((((.......)))))))(((.(((....))))))))..)((....))))))))))..... ( -34.50) >consensus GGAUUGGGGGUCUG_GU_________CUGGGCUGCCUGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUU ((((.....))))........................((....((((((((((((.......)))))))(((((((....))))).)))))))....))............. (-25.66 = -26.05 + 0.39)
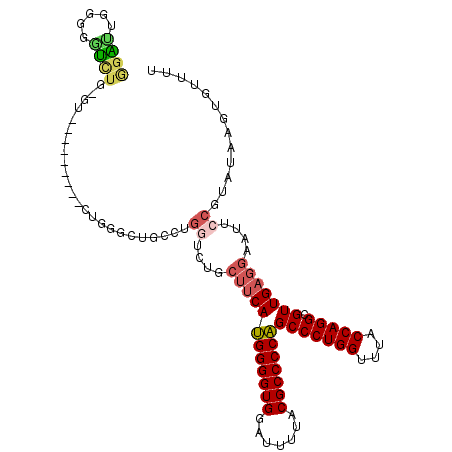
| Location | 3,845,542 – 3,845,644 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -18.04 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3845542 102 - 20766785 AAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCAGGCAGCUCAG---------AC-CAGACCCCCAAUCC .............((....)).....(((((((....(((((.((((........)))).))).))......))))))).......---------..-.............. ( -24.70) >DroSec_CAF1 101474 97 - 1 AAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAGUCCACCCCAUGAAGCAGAACAGGCAGCCCAG---------------ACCCACAAUCC .............((..............(((....)))((((((((((.....))))...((.((........))))))))))..---------------..))....... ( -22.70) >DroSim_CAF1 118936 103 - 1 AAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCUGGCAGCCCAG---------ACCCAGACCCACAAUCC .............(((.............(((....)))((((((((((...................))....((((....))))---------.))))).)))....))) ( -24.31) >DroEre_CAF1 102767 109 - 1 AAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAGCCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCGGACCAGGCAGCCCAGGCAGCCCAGAC-CAGA-C-CAAGCCC .............((...........(((((((..(((((((.((((........)))).))).))..))..))))))).((....))........)-)...-.-....... ( -32.00) >DroYak_CAF1 99748 92 - 1 GAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGC------------------AACCCAGGC-CAAACC-CAAGCCC .............((............(((((....)))))(((((((.(.......).))))....)))------------------...)).(((-......-...))). ( -21.50) >DroAna_CAF1 97270 96 - 1 GAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCAGGGGCGUUAAAUCCACCCCUUG------GCCAAGCAGCAAGU---------UC-CAGGUCCAGGACCU .............((((((.........((((....))))((((((((.(.......).)))))..------)))........)))---------))-)(((((...))))) ( -28.00) >consensus AAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCAGGCAGCCCAG_________AC_CAGACCCACAACCC .............((....))(((...(((((....)))))..(((((.(.......).))))))))............................................. (-18.04 = -17.93 + -0.11)

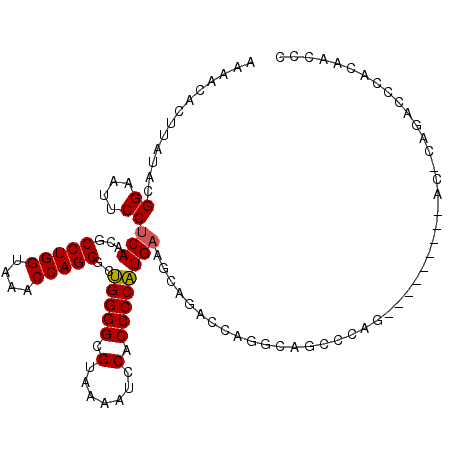
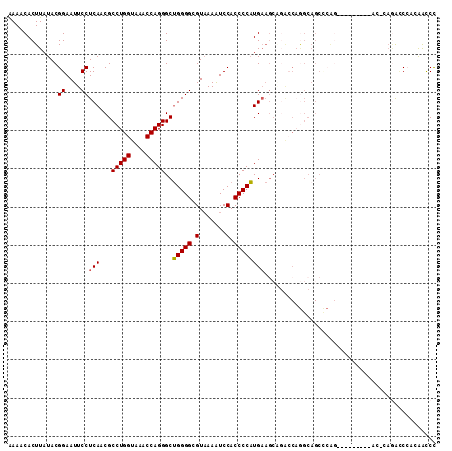
| Location | 3,845,564 – 3,845,675 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
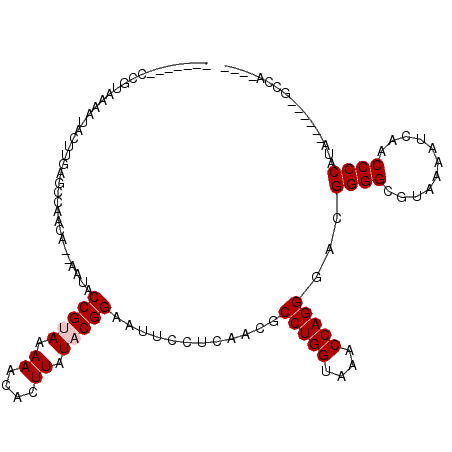
>2R_DroMel_CAF1 3845564 111 + 20766785 UGCCUGGUCUGCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUU--UGUUGGCUCAAGCAUUUUACGG------- .(((((((..(((...(((((((.......))))))).....)))..)))))))((..(.((((.(((((..........))))).)))--).)..))...............------- ( -43.90) >DroPse_CAF1 94662 103 + 1 ----GGGC------UCUGGGGUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGCACAAGUAUUUUUCCG------- ----((((------((.(((((.........)))))(.((((((....))))).).)))).............(((((...((((.....)))).))))).........))).------- ( -30.20) >DroWil_CAF1 156340 113 + 1 GGCAAAAGCUUUUUUUUGGGGUUGAUUUUACGCCCCGUCCCUGGCUUACCAGGCGUUGAGAAAUUCCGUA-AAGUGUUUUUACGGUAUU--UUU-G---CCAGUGUUAUACGACGACGAC ((((((((...(((((((((((.........)))))...(((((....)))))....))))))..(((((-(((...))))))))..))--)))-)---)).(((((....))).))... ( -32.10) >DroYak_CAF1 99770 101 + 1 ----------GCUUCAUGGGGUGGAUUUUACGCCCCAGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUU--UGUUGGCUCAAGCAUUUUACGG------- ----------((((..(((((((.......)))))))(((.(((....))))))((..(.((((.(((((..........))))).)))--).)..))..)))).........------- ( -37.10) >DroAna_CAF1 97292 105 + 1 UGCUUGGC------CAAGGGGUGGAUUUAACGCCCCUGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUCUACGGUAUU--UGUUGGCUCAAGCAUUUUCCGG------- ((((((((------(.(((((((.......)))))))(((((((....))))).)).((.((((.(((((..........))))).)))--).))))).))))))........------- ( -42.70) >DroPer_CAF1 95580 103 + 1 ----GGGC------UCUGGGGUUGAUUUAACGCCCCGUCCCUGGUCUACCAGGCGUCGAGGAAUUCCGAAAAAGUGUUUUUACGGUAUUUCUGUUGGCACAAGUAUUUUUCCG------- ----((((------((.(((((.........)))))(.((((((....))))).).)))).............(((((...((((.....)))).))))).........))).------- ( -30.20) >consensus ____GGGC______CAUGGGGUGGAUUUAACGCCCCGGCCCUGGUUUACCAGGCGUUGAGGAAUUCCGUAUAAGUGUUUUUACGGUAUU__UGUUGGCUCAAGCAUUUUACGG_______ ................((((((.........))))))..(((((....))))).(((((.((((.(((((..........))))).)))..).)))))...................... (-21.07 = -21.10 + 0.03)

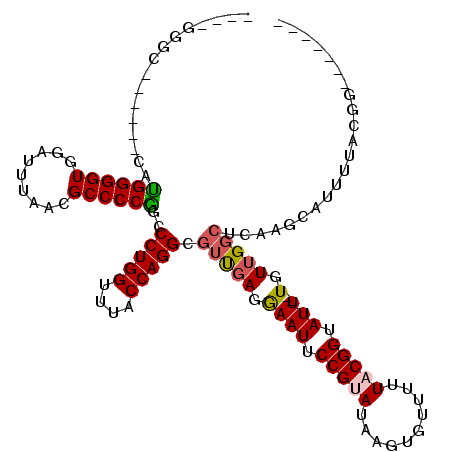
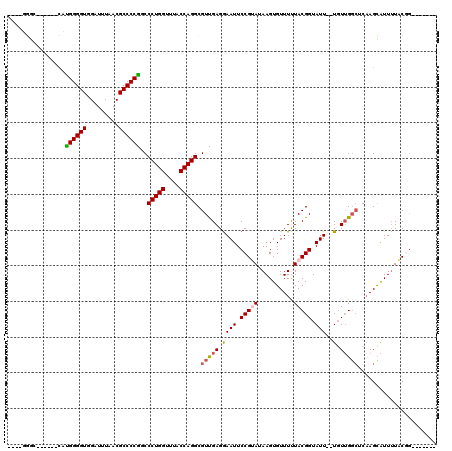
| Location | 3,845,564 – 3,845,675 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.05 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3845564 111 - 20766785 -------CCGUAAAAUGCUUGAGCCAACA--AAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGCAGACCAGGCA -------........((((((((((....--....(((((.((....)).))))).............((((....))))))))((((.(.......).))))...........)))))) ( -30.80) >DroPse_CAF1 94662 103 - 1 -------CGGAAAAAUACUUGUGCCAACAGAAAUACCGUAAAAACACUUUUUCGGAAUUCCUCGACGCCUGGUAGACCAGGGACGGGGCGUUAAAUCAACCCCAGA------GCCC---- -------.((((......((((....)))).....(((.((((....)))).)))..)))).((.(.(((((....)))))).))(((.(((.....))))))...------....---- ( -24.20) >DroWil_CAF1 156340 113 - 1 GUCGUCGUCGUAUAACACUGG---C-AAA--AAUACCGUAAAAACACUU-UACGGAAUUUCUCAACGCCUGGUAAGCCAGGGACGGGGCGUAAAAUCAACCCCAAAAAAAAGCUUUUGCC ...................((---(-(((--(...(((((((.....))-)))))............(((((....)))))...((((...........))))..........))))))) ( -29.70) >DroYak_CAF1 99770 101 - 1 -------CCGUAAAAUGCUUGAGCCAACA--AAUACCGUAGAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCUGGGGCGUAAAAUCCACCCCAUGAAGC---------- -------.........((((.((((....--....(((((..........))))).............((((....))))))))((((.(.......).))))...))))---------- ( -26.80) >DroAna_CAF1 97292 105 - 1 -------CCGGAAAAUGCUUGAGCCAACA--AAUACCGUAGAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGCAGGGGCGUUAAAUCCACCCCUUG------GCCAAGCA -------........((((((.((((...--....(((((..........)))))...........((((((....))).)))(((((.(.......).)))))))------)))))))) ( -35.70) >DroPer_CAF1 95580 103 - 1 -------CGGAAAAAUACUUGUGCCAACAGAAAUACCGUAAAAACACUUUUUCGGAAUUCCUCGACGCCUGGUAGACCAGGGACGGGGCGUUAAAUCAACCCCAGA------GCCC---- -------.((((......((((....)))).....(((.((((....)))).)))..)))).((.(.(((((....)))))).))(((.(((.....))))))...------....---- ( -24.20) >consensus _______CCGUAAAAUACUUGAGCCAACA__AAUACCGUAAAAACACUUAUACGGAAUUCCUCAACGCCUGGUAAACCAGGGACGGGGCGUAAAAUCAACCCCAUA______GCCA____ ...................................(((((.((....)).)))))............(((((....)))))...((((...........))))................. (-18.25 = -18.58 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:06 2006