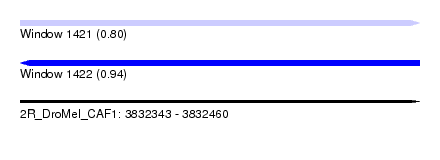
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,832,343 – 3,832,460 |
| Length | 117 |
| Max. P | 0.939750 |

| Location | 3,832,343 – 3,832,460 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -22.86 |
| Energy contribution | -22.42 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3832343 117 + 20766785 UUAACUUUUCCUCUCAGCUCAUGUCCACAUUCACAUAUUCUUGGUGAACCCAAUGGGGGUUGCCAAUGCUUUUUAAGAUCCGAAGGGAUUGGGCGGCUGCUACCGCCCUAAUCCUUG ...............(((..((((........))))....(((((.(((((.....)))))))))).))).............(((((((((((((......)))))).))))))). ( -34.70) >DroSec_CAF1 88420 117 + 1 UUAACUUUUUCUCUCAGCUCAUGACCGCAUUCAGAUAUUCUUGGUGAACCCAAUGUGGGUUGGCAAUGCUUUUUAAGAUCCGAGGGGAUUGGGCGGCUGCUACCGCCCUAAUCCUUG ............(((.(.((.(((..((((((((......)))((.(((((.....))))).)))))))...))).)).).)))((((((((((((......)))))).)))))).. ( -37.00) >DroSim_CAF1 106109 117 + 1 UUAACUUUUCCUCUCAGCUCAUGACUACAUUCAGAUAUUCUUGGUGAACCCAAUGCGGAUUGGCAAUGCUUUUUAAGAUCCGAGGGGAUUGGGCGGCUGCUACCGCCCUAAUCCUUG .........((((...((((.(((......))))).......((....))....))((((((((...)))......)))))))))(((((((((((......)))))).)))))... ( -33.20) >DroEre_CAF1 90156 99 + 1 UUAUCUUUUUCUCUC--------------U----UUAUUCUUGACGAAACCAAUGCGGUUUUCCAACGUUUCUUAAGAUGCGCAGGGAUUGGGCGGCUGCUCCCGCCCUAAUCCUUG ...............--------------.----....((((((.(((((......((....))...)))))))))))....((((((((((((((......)))))).)))))))) ( -30.00) >DroYak_CAF1 86843 117 + 1 UUAACUUUUUCUCUUAGCUCAUGUCCACGUUUAGAUAUUCUCGGCGAACACAACGCGGUUUUCCAAUGCUUUUUGAAAUCAGAAGAGAUUGGGCGGCUGCUUCCGCCCUAAUCCUUG ....((.((((....(((...(((...(((..((.....))..)))...)))....((....))...)))....))))..))(((.((((((((((......)))))).))))))). ( -28.00) >consensus UUAACUUUUUCUCUCAGCUCAUGACCACAUUCAGAUAUUCUUGGUGAACCCAAUGCGGGUUGCCAAUGCUUUUUAAGAUCCGAAGGGAUUGGGCGGCUGCUACCGCCCUAAUCCUUG ......................................(((((((((((((.....))))).....))))....)))).....(((((((((((((......)))))).))))))). (-22.86 = -22.42 + -0.44)


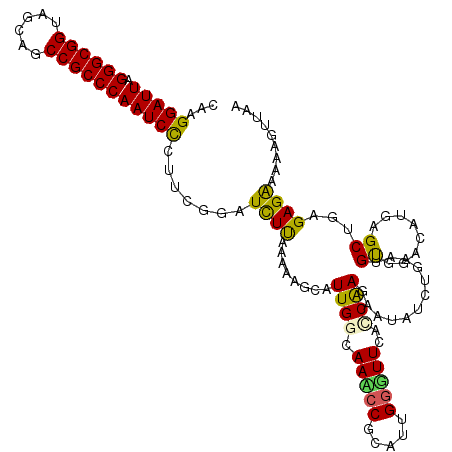
| Location | 3,832,343 – 3,832,460 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -23.24 |
| Energy contribution | -22.74 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3832343 117 - 20766785 CAAGGAUUAGGGCGGUAGCAGCCGCCCAAUCCCUUCGGAUCUUAAAAAGCAUUGGCAACCCCCAUUGGGUUCACCAAGAAUAUGUGAAUGUGGACAUGAGCUGAGAGGAAAAGUUAA ...(((((.(((((((....))))))))))))(....).((((....(((.((((.(((((.....)))))..))))...(((((........))))).)))...))))........ ( -38.20) >DroSec_CAF1 88420 117 - 1 CAAGGAUUAGGGCGGUAGCAGCCGCCCAAUCCCCUCGGAUCUUAAAAAGCAUUGCCAACCCACAUUGGGUUCACCAAGAAUAUCUGAAUGCGGUCAUGAGCUGAGAGAAAAAGUUAA ...(((((.(((((((....)))))))))))).(((((.((.......(((((...(((((.....))))).....((.....)).)))))......)).)))))............ ( -37.72) >DroSim_CAF1 106109 117 - 1 CAAGGAUUAGGGCGGUAGCAGCCGCCCAAUCCCCUCGGAUCUUAAAAAGCAUUGCCAAUCCGCAUUGGGUUCACCAAGAAUAUCUGAAUGUAGUCAUGAGCUGAGAGGAAAAGUUAA ...(((((.(((((((....))))))))))))((((....((((....(((((.(((((....)))))((((.....)))).....))))).....))))....))))......... ( -35.40) >DroEre_CAF1 90156 99 - 1 CAAGGAUUAGGGCGGGAGCAGCCGCCCAAUCCCUGCGCAUCUUAAGAAACGUUGGAAAACCGCAUUGGUUUCGUCAAGAAUAA----A--------------GAGAGAAAAAGAUAA ...(((((.((((((......)))))))))))((.....((((........((((.(((((.....)))))..))))......----.--------------)))).....)).... ( -25.66) >DroYak_CAF1 86843 117 - 1 CAAGGAUUAGGGCGGAAGCAGCCGCCCAAUCUCUUCUGAUUUCAAAAAGCAUUGGAAAACCGCGUUGUGUUCGCCGAGAAUAUCUAAACGUGGACAUGAGCUAAGAGAAAAAGUUAA .........((((((......))))))..((((((....((((((......))))))..(((((((((((((.....))))))...))))))).........))))))......... ( -35.20) >consensus CAAGGAUUAGGGCGGUAGCAGCCGCCCAAUCCCUUCGGAUCUUAAAAAGCAUUGGCAAACCGCAUUGGGUUCACCAAGAAUAUCUGAAUGUGGACAUGAGCUGAGAGAAAAAGUUAA ...(((((.((((((......))))))))))).......((((........((((.(((((.....)))))..))))............((........))...))))......... (-23.24 = -22.74 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:48:02 2006