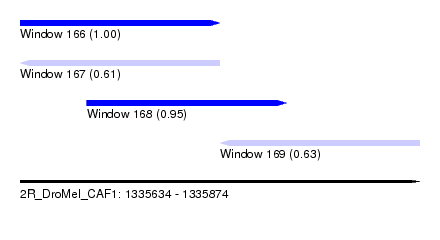
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,335,634 – 1,335,874 |
| Length | 240 |
| Max. P | 0.998272 |

| Location | 1,335,634 – 1,335,754 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1335634 120 + 20766785 CACAUCGUUUGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGC (((....((((((((((((.....))))).))))))).((((((((.........(((((((.((((((((.......)))))))))))))))..))))))))))).............. ( -35.80) >DroSec_CAF1 54155 120 + 1 CACGUCGUUUGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCCGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGC ((((((.((((((((((((.....))))).)))))))))).....(((.(((...(((((((.((((((((.......)))))))))))))))..)))..)))))).............. ( -40.80) >DroSim_CAF1 32345 120 + 1 CACAUCGUUUGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGC (((....((((((((((((.....))))).))))))).((((((((.........(((((((.((((((((.......)))))))))))))))..))))))))))).............. ( -35.80) >DroEre_CAF1 35688 120 + 1 CACAUCGUUAGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCCGGCGAUUAUGGCAAAUGAAGCGAAACUGUUUGUGUGAUUCCUUGUUUGGA ....((((.(((.........)))))))((.((((((((...((.((((....(((((((((.((((((((.......)))))))))))))))..))....)))).)))))))))).)). ( -36.00) >DroYak_CAF1 22844 120 + 1 CACAUCGUUAGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGCCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUCUUGUUUAGA (((.......(((((((((.....))))).))))....((((((((.........(((((((.(((((((.........))))))))))))))..))))))))))).............. ( -33.60) >consensus CACAUCGUUUGUCAAUCGUAAACUGCGAUCUGACAGGGACAAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGC (((....((((((((((((.....))))).))))))).((((((((.........(((((((.((((((((.......)))))))))))))))..))))))))))).............. (-35.14 = -35.14 + 0.00)

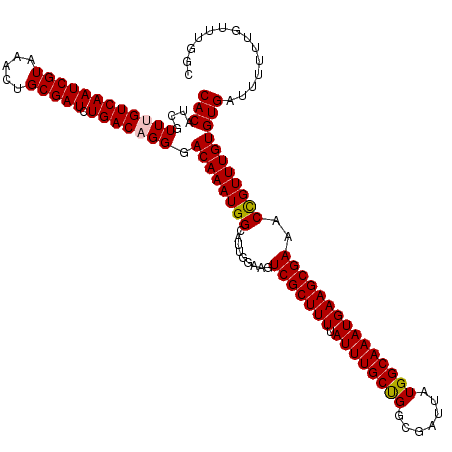
| Location | 1,335,634 – 1,335,754 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1335634 120 - 20766785 GCCAAACAAAAAAUCACACAAACGGUUUCGCUUCAUUUGCCAUAAUCGCCAGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACAAACGAUGUG ................((((...((..((((((.((((((...........))))))..))))))...))........((((...((((((.(((.......))))))))).)))))))) ( -24.20) >DroSec_CAF1 54155 120 - 1 GCCAAACAAAAAAUCACACAAACGGUUUCGCUUCAUUUGCCAUAAUCGCCGGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACAAACGACGUG ................(((....((..((((((.(((((((.........)))))))..))))))...))..........(((..((((((.(((.......)))))))))...)))))) ( -29.70) >DroSim_CAF1 32345 120 - 1 GCCAAACAAAAAAUCACACAAACGGUUUCGCUUCAUUUGCCAUAAUCGCCAGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACAAACGAUGUG ................((((...((..((((((.((((((...........))))))..))))))...))........((((...((((((.(((.......))))))))).)))))))) ( -24.20) >DroEre_CAF1 35688 120 - 1 UCCAAACAAGGAAUCACACAAACAGUUUCGCUUCAUUUGCCAUAAUCGCCGGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACUAACGAUGUG (((......)))....((((..(((((((((((.(((((((.........)))))))..)))))).......(((.(((((.......))))).)))......))))).(....).)))) ( -26.80) >DroYak_CAF1 22844 120 - 1 UCUAAACAAGAAAUCACACAAACGGUUUCGCUUCAUUUGCCAUAAUCGGCAGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACUAACGAUGUG ................((((...((..((((((...(((((......))))).......))))))...))........((((....(((((.(((.......))))))))..)))))))) ( -23.80) >consensus GCCAAACAAAAAAUCACACAAACGGUUUCGCUUCAUUUGCCAUAAUCGCCAGCAAAUAAAAGCGACUUCCAAUGCCAUUUGUCCCUGUCAGAUCGCAGUUUACGAUUGACAAACGAUGUG ..............((((((((.(((.((((((.((((((...........))))))..))))))........))).)))))....(((((.(((.......)))))))).......))) (-22.54 = -22.74 + 0.20)

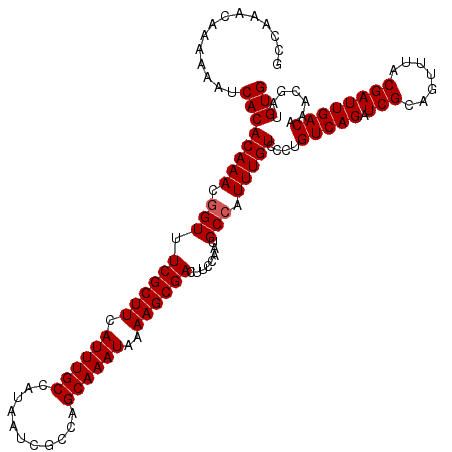
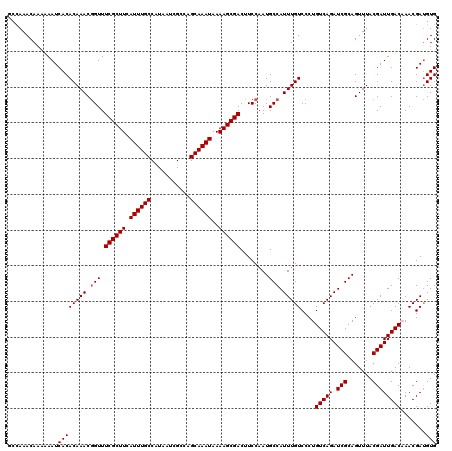
| Location | 1,335,674 – 1,335,794 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -31.06 |
| Energy contribution | -32.10 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1335674 120 + 20766785 AAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGCCAGCGCGAAGGACUCGUGCUACCAGACAUUUUUGCGGACA ..........((...(((((((.((((((((.......)))))))))))))))..))(((((((.((.....(((((((..(((((((.....))))))).))))))).)).))))))). ( -41.40) >DroSec_CAF1 54195 120 + 1 AAAUGGCAUUGGAAGUCGCUUUUAUUUGCCGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGCCAGCGCGAAGGACUCCUGCUGCCGGACAUUUGUGCGGACA ..........((...(((((((.((((((((.......)))))))))))))))..))(((((((..(.....((((((((.((((.((.....)).)))))))))))).)..))))))). ( -44.60) >DroSim_CAF1 32385 120 + 1 AAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGCCAGCGCGAAGGACUCCUGCUGCCGGACAUUUUUGCGGACA ..........((...(((((((.((((((((.......)))))))))))))))..))(((((((.((.....((((((((.((((.((.....)).)))))))))))).)).))))))). ( -38.80) >DroEre_CAF1 35728 117 + 1 AAAUGGCAUUGGAAGUCGCUUUUAUUUGCCGGCGAUUAUGGCAAAUGAAGCGAAACUGUUUGUGUGAUUCCUUGUUUGGACAGCGCGAAGGACAACUGCUGCGAGAAGUUUUUGCAG--- ....(((((((....(((((((.((((((((.......)))))))))))))))...(.(((((((...(((......)))..))))))).).))).))))((((((...))))))..--- ( -37.80) >DroYak_CAF1 22884 120 + 1 AAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGCCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUCUUGUUUAGACAACGCGAACGACUCCUGCCGCUCGAAAUGUUUGCGGACA ....((((..(((..(((((((.(((((((.........))))))))))))))...(((((((((....(((.....)))..)))))))))..)))))))((...........))..... ( -36.60) >consensus AAAUGGCAUUGGAAGUCGCUUUUAUUUGCUGGCGAUUAUGGCAAAUGAAGCGAAACCGUUUGUGUGAUUUUUUGUUUGGCCAGCGCGAAGGACUCCUGCUGCCAGACAUUUUUGCGGACA ((((.(((((((...(((((((.((((((((.......)))))))))))))))..)))...)))).))))..(((((((.(((((.((.....)).))))))))))))............ (-31.06 = -32.10 + 1.04)


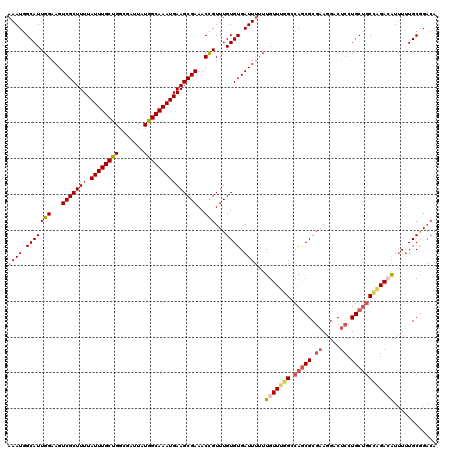
| Location | 1,335,754 – 1,335,874 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.23 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -31.55 |
| Energy contribution | -31.99 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1335754 120 - 20766785 AUCAGCCCUGAUUACAUUAGACAUUAAAAGCAGGGAAGCGAGUCAGUGCUUGGCGGUCAGCUGAGGCCACUUCGGUUGGCUGUCCGCAAAAAUGUCUGGUAGCACGAGUCCUUCGCGCUG ((((....))))...((((((((((..(((((..((......))..)))))(((((((((((((((...)))))))))))))))......))))))))))(((.((((...)))).))). ( -42.80) >DroSec_CAF1 54275 115 - 1 AUCAGCCCUGAUUACAUUAGACAUUAAAAGCAGGG-----AGUCAGUGCUGGGCGGUCAGCCGAGGGCACUUCGGUUGGCUGUCCGCACAAAUGUCCGGCAGCAGGAGUCCUUCGCGCUG ..((((.(((((...))))).........((((((-----(.((.((((.((((((((((((((((...))))))))))))))))))))...(((......))).)).))))).)))))) ( -50.10) >DroSim_CAF1 32465 114 - 1 AUCA-CCCUGAUUACAUUAGACAUUAAAAGCAGGG-----AGUCAGUGCUGGGCGGUCAGCCGAGGGCACUUCGGUUGGCUGUCCGCAAAAAUGUCCGGCAGCAGGAGUCCUUCGCGCUG ....-..(((((...))))).........((((((-----(.((..((((((((((((((((((((...))))))))))))))))((...........)))))).)).))))).)).... ( -44.40) >DroYak_CAF1 22964 115 - 1 AUCAGCCCUAAUUACAUUAGGCAUUUAAAGCAGGG-----AGUCAGUGCUUGGCUGUUAGCUGCGGCCACUUUGGUUGGCUGUCCGCAAACAUUUCGAGCGGCAGGAGUCGUUCGCGUUG ..((((((((((...))))))........((.((.-----(((((..((((((((((.....)))))))....)))))))).)).))........((((((((....)))))))).)))) ( -36.50) >consensus AUCAGCCCUGAUUACAUUAGACAUUAAAAGCAGGG_____AGUCAGUGCUGGGCGGUCAGCCGAGGCCACUUCGGUUGGCUGUCCGCAAAAAUGUCCGGCAGCAGGAGUCCUUCGCGCUG .....(((((....................)))))........((((((.((((((((((((((((...))))))))))))))))((..............))...........)))))) (-31.55 = -31.99 + 0.44)

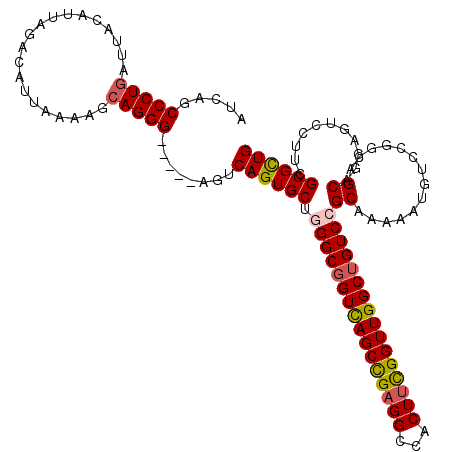
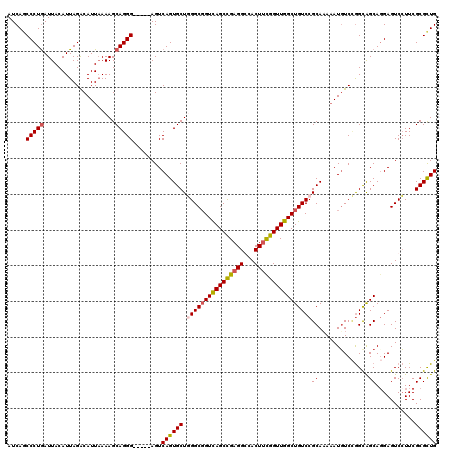
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:14 2006