
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,808,820 – 3,808,925 |
| Length | 105 |
| Max. P | 0.992484 |

| Location | 3,808,820 – 3,808,925 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
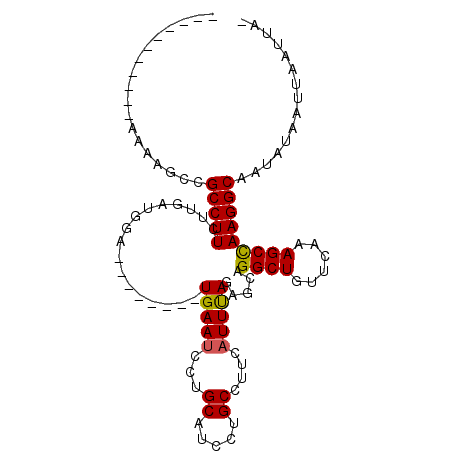
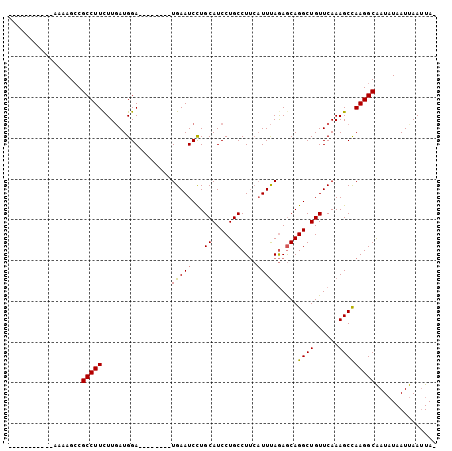
>2R_DroMel_CAF1 3808820 105 + 20766785 UUUGCAAAAUGAAAAGACGCCUUCUUGAUGGAUAUUCAGCUGAAUCCUGCAUCCUGCCUGAUUUUAAAACAGGCUGUUCAAAGCUAAGGCGAUCUAAUCAAAUAU .........(((..(((((((((..(..((((((....((........)).....(((((.........)))))))))))..)..)))))).)))..)))..... ( -24.80) >DroSec_CAF1 65265 85 + 1 -----------AAAAGCCGCCUUUUUGAUGGA--------UGAAUCUUGCAUCCUGCCUUCAUUUAGAGCAGGCUGUUCAAAGCCAAGGCAAUAUAAUUAAUUA- -----------....(((((((...(((.(((--------((.......))))).....)))...)).)).((((......))))..)))..............- ( -23.10) >DroSim_CAF1 78845 84 + 1 -----------AA-AGCCGCCUUCUUUAUGGA--------UGAAUCCUGCAUCCUGCCUUCAUUCAGAGCAGGCUGUUCAAAGCCAAGGCAAUAUAAUUAAUUA- -----------..-....(((((.....((((--------((((....((.....)).)))))))).....((((......)))))))))..............- ( -21.30) >consensus ___________AAAAGCCGCCUUCUUGAUGGA________UGAAUCCUGCAUCCUGCCUUCAUUUAGAGCAGGCUGUUCAAAGCCAAGGCAAUAUAAUUAAUUA_ ..................(((((.................(((((...((.....))....))))).....((((......)))))))))............... (-14.04 = -13.93 + -0.11)

| Location | 3,808,820 – 3,808,925 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.71 |
| Energy contribution | -15.50 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
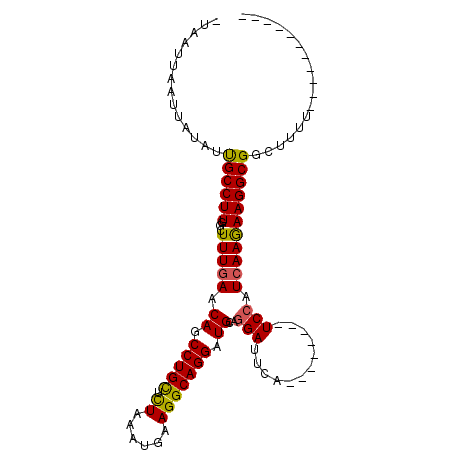
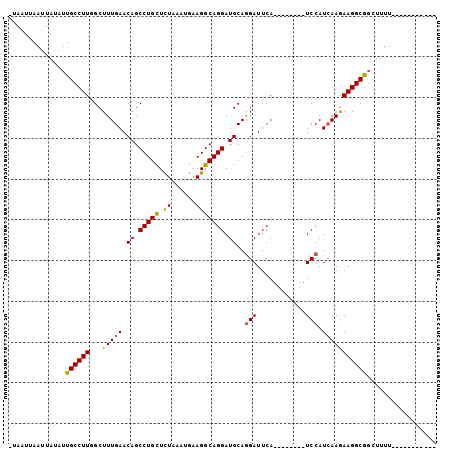
>2R_DroMel_CAF1 3808820 105 - 20766785 AUAUUUGAUUAGAUCGCCUUAGCUUUGAACAGCCUGUUUUAAAAUCAGGCAGGAUGCAGGAUUCAGCUGAAUAUCCAUCAAGAAGGCGUCUUUUCAUUUUGCAAA .....(((..(((.((((((...(((((...(((((.........))))).((((((((.......)))..))))).))))))))))))))..)))......... ( -30.80) >DroSec_CAF1 65265 85 - 1 -UAAUUAAUUAUAUUGCCUUGGCUUUGAACAGCCUGCUCUAAAUGAAGGCAGGAUGCAAGAUUCA--------UCCAUCAAAAAGGCGGCUUUU----------- -............(((((((...(((((...((((.(.......).)))).(((((.......))--------))).)))))))))))).....----------- ( -21.10) >DroSim_CAF1 78845 84 - 1 -UAAUUAAUUAUAUUGCCUUGGCUUUGAACAGCCUGCUCUGAAUGAAGGCAGGAUGCAGGAUUCA--------UCCAUAAAGAAGGCGGCU-UU----------- -............((((((...(((((..((.(((((.((......))))))).))..(((....--------))).))))).))))))..-..----------- ( -22.30) >consensus _UAAUUAAUUAUAUUGCCUUGGCUUUGAACAGCCUGCUCUAAAUGAAGGCAGGAUGCAGGAUUCA________UCCAUCAAGAAGGCGGCUUUU___________ ..............((((((...(((((.((.(((((.((......))))))).))..(((............))).)))))))))))................. (-15.71 = -15.50 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:47 2006