
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,802,617 – 3,802,717 |
| Length | 100 |
| Max. P | 0.892574 |

| Location | 3,802,617 – 3,802,717 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -34.32 |
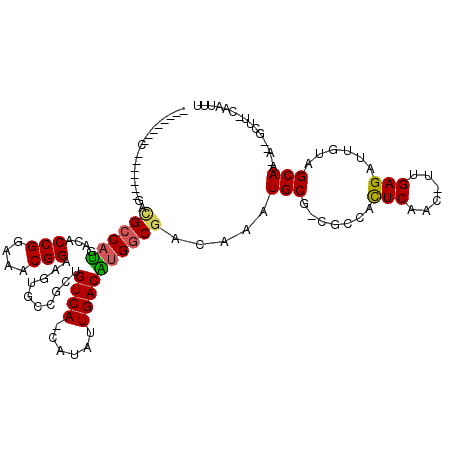
| Consensus MFE | -21.50 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
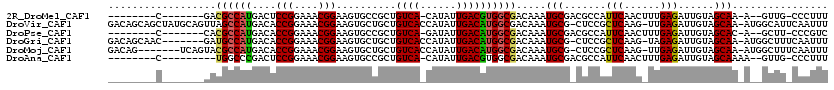
>2R_DroMel_CAF1 3802617 100 + 20766785 --------C-------GACGCCAUGACUCCGGAAACGGAAGUGCCGCUGUCA-CAUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAA-A--GUUG-CCCUUU --------(-------(.(((......((((....)))).(((((((.((((-.....))))))))).))....))).)).....((((((((.........)))-)--))))-...... ( -31.80) >DroVir_CAF1 68592 117 + 1 GACAGCAGCUAUGCAGUUAGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAG-UUGAGAUUGUAGCAA-AUGGCAUUCAAUUU (((.(((....))).))).(((((....(((....)))...((((((((((.((((......)))).))))......-(((.((....)-).)))...)))))).-)))))......... ( -38.80) >DroPse_CAF1 55163 100 + 1 --------C-------CACGCCAUGACACCGGAAACGGAAGUGCCGCUGUCA-GAUAUUGACAUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAC-A--GCUU-CCCGUC --------.-------........(((.(((....)))..((((..(.(((.-((..((((.((((((.(......).))))))))))..))..))).)..))))-.--....-...))) ( -31.50) >DroGri_CAF1 60340 110 + 1 GACAGCAAC-------GAUGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAG-UAGAGAUUGUAGCAA-AUGGCUUUCAAUUU .........-------((.(((((....(((....)))...((((((((((.((((......)))).))))......-(((..(....)-..)))...)))))).-)))))..))..... ( -36.20) >DroMoj_CAF1 83483 110 + 1 GACAG-------UCAGUACGCCAUGACACCGGAAACGGAAGUGCUGCUGUCACCAUAUUGACAUGGCGACAAAUGCG-CUCCGCUCAAG-UUGAGAUUGUAGCAA-AUGGCUUUCAAUUU ((.((-------(((((.(((((((...(((....)))..((((....).)))........)))))))))...(((.-.....(((...-..)))......))).-.))))).))..... ( -35.40) >DroAna_CAF1 56017 99 + 1 --------C---------UGGCCCGACUCCGGAAACGGAAGUGCCGCUGUCA-CAUAUUGACGUGGCGACAAAUGCGACGCCAUUCAACUUUGAGAUUGUAGCAAAA--GUUG-CCCUUU --------.---------.(((..(((((((....))))..(((..(.(((.-((..((((.((((((.(......).))))))))))...)).))).)..)))...--))))-)).... ( -32.20) >consensus ________C_______GACGCCAUGACACCGGAAACGGAAGUGCCGCUGUCA_CAUAUUGACAUGGCGACAAAUGCG_CGCCACUCAAC_UUGAGAUUGUAGCAA_A__GCUU_CAAUUU ..................((((((....(((....)))..........((((......)))))))))).....(((.......(((......)))......)))................ (-21.50 = -21.22 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:45 2006