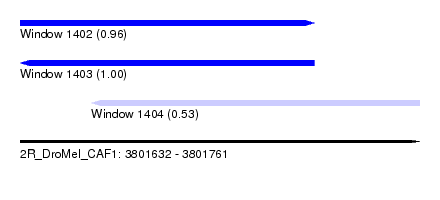
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,801,632 – 3,801,761 |
| Length | 129 |
| Max. P | 0.999866 |

| Location | 3,801,632 – 3,801,727 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.93 |
| Mean single sequence MFE | -18.12 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
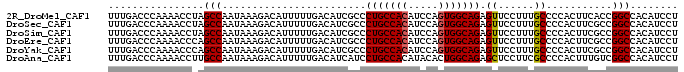
>2R_DroMel_CAF1 3801632 95 + 20766785 UUUGACCCAAAACCUAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCACCGGCCACAUCCU ................(((........((....)).....((.(((((((.....))))))).))...................)))........ ( -17.50) >DroSec_CAF1 58037 95 + 1 UUUGACCCAAAACCUAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCGCCGGCCACAUCCU ................(((...((((.((....)).....((.(((((((.....))))))).))..))))((........)).)))........ ( -18.10) >DroSim_CAF1 71596 95 + 1 UUUGACCCAAAACCUAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCGCCGGCCACAUCCU ................(((...((((.((....)).....((.(((((((.....))))))).))..))))((........)).)))........ ( -18.10) >DroEre_CAF1 59401 95 + 1 UUUGACCCAAAACCCAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCGCCGGCCACAUCCU ................(((...((((.((....)).....((.(((((((.....))))))).))..))))((........)).)))........ ( -18.10) >DroYak_CAF1 54988 95 + 1 UUUGACCCAAAACCCAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCGCCGGCCACAUCCU ................(((...((((.((....)).....((.(((((((.....))))))).))..))))((........)).)))........ ( -18.10) >DroAna_CAF1 55143 95 + 1 UUUGACCCAAAACCUUGCCAAUAAAGACAUUUUUGACAUCAUCCUGCCACAUACACUGGCAGAGCUCCUUCGCCCCACUUUGUCGGCCACAUCCU ................(((.((((((.................((((((.......)))))).((......))....)))))).)))........ ( -18.80) >consensus UUUGACCCAAAACCUAGCCAAUAAAGACAUUUUUGACAUCGCCCUGCCACAUCCAGUGGCAGAGUUCCUUUGCCCCACUUCGCCGGCCACAUCCU ................(((........................(((((((.....))))))).((......))...........)))........ (-16.07 = -16.10 + 0.03)
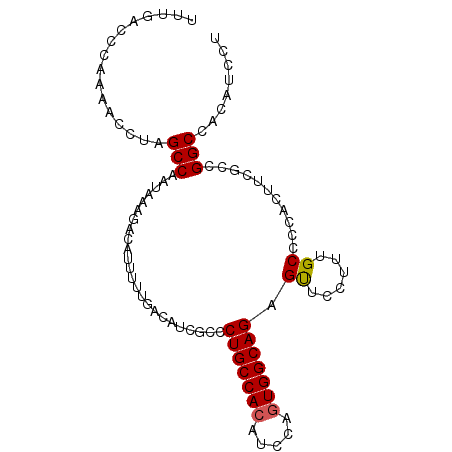
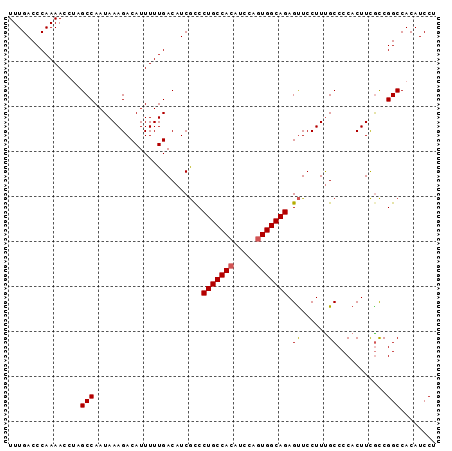
| Location | 3,801,632 – 3,801,727 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 95.93 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.17 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3801632 95 - 20766785 AGGAUGUGGCCGGUGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUAGGUUUUGGGUCAAA (((((.((((((((((((...((((..(...(((((((((.....))))))))))..))))........)))))))))))).)))))........ ( -36.60) >DroSec_CAF1 58037 95 - 1 AGGAUGUGGCCGGCGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUAGGUUUUGGGUCAAA (((((.(((((((..(((...((((..(...(((((((((.....))))))))))..))))........)))..))))))).)))))........ ( -33.20) >DroSim_CAF1 71596 95 - 1 AGGAUGUGGCCGGCGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUAGGUUUUGGGUCAAA (((((.(((((((..(((...((((..(...(((((((((.....))))))))))..))))........)))..))))))).)))))........ ( -33.20) >DroEre_CAF1 59401 95 - 1 AGGAUGUGGCCGGCGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUGGGUUUUGGGUCAAA ..(((.((.(((((.((..((((((...((.(((((((((.....))))))))).....)).....))))))..)).))))).)...).)))... ( -32.10) >DroYak_CAF1 54988 95 - 1 AGGAUGUGGCCGGCGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUGGGUUUUGGGUCAAA ..(((.((.(((((.((..((((((...((.(((((((((.....))))))))).....)).....))))))..)).))))).)...).)))... ( -32.10) >DroAna_CAF1 55143 95 - 1 AGGAUGUGGCCGACAAAGUGGGGCGAAGGAGCUCUGCCAGUGUAUGUGGCAGGAUGAUGUCAAAAAUGUCUUUAUUGGCAAGGUUUUGGGUCAAA ......(((((.(((..(..((((......))))..)...)))......((((((..((((((.((.....)).))))))..))))))))))).. ( -27.30) >consensus AGGAUGUGGCCGGCGAAGUGGGGCAAAGGAACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUUUAUUGGCUAGGUUUUGGGUCAAA (((((.(((((((.((((...((((..(...(((((((((.....))))))))))..))))........)))).))))))).)))))........ (-30.44 = -30.17 + -0.28)


| Location | 3,801,655 – 3,801,761 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -19.88 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3801655 106 - 20766785 GCAAGACGACGCGCAGACAAAAUGUUUGCUAGACAGGAUGUGGCCGGUGAAGUGGGG-CAAAGG---AACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUU ((........))((((((.....)))))).(((((.(((((.(((........((..-(....)---..))(((((((.....)))))))))).))))).....))))). ( -36.70) >DroPse_CAF1 53945 105 - 1 ACAAGACGACGCGCAGACGAAAUAUUUGUUAGACAGGAAACGG-----GAAGUGGAGUCGAAGGGCGGACACUGCCAUCGAACCGGCUGGAAAGAUGUCAAAAAUGUCUC ...(((((.......(((((.....))))).(((((....)..-----..(((.(..((((..(((((...))))).))))..).))).......)))).....))))). ( -26.50) >DroEre_CAF1 59424 106 - 1 GCAAGACGACGCGCAGACAAAAUGUUUGCUAGACAGGAUGUGGCCGGCGAAGUGGGG-CAAAGG---AACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUU ..((((((....((((((.....))))))..((((.....((.((.((...)).)).-))....---..(((((((((.....)))))))))...)))).....)))))) ( -38.60) >DroYak_CAF1 55011 106 - 1 GCAAGACGACGCGCAGACAAAAUGUUUGCUAGACAGGAUGUGGCCGGCGAAGUGGGG-CAAAGG---AACUCUGCCACUGGAUGUGGCAGGGCGAUGUCAAAAAUGUCUU ..((((((....((((((.....))))))..((((.....((.((.((...)).)).-))....---..(((((((((.....)))))))))...)))).....)))))) ( -38.60) >DroAna_CAF1 55166 106 - 1 GCAAGACGACGCGCAGACAAAAUGUUUGCUAGACAGGAUGUGGCCGACAAAGUGGGG-CGAAGG---AGCUCUGCCAGUGUAUGUGGCAGGAUGAUGUCAAAAAUGUCUU ..((((((....((((((.....))))))..((((..((.(.(((((((..(..(((-(.....---.))))..)...)))...)))).).))..)))).....)))))) ( -34.70) >DroPer_CAF1 53097 105 - 1 ACAAGACGACGCGCAGACGAAAUAUUUGUUAGACAGGAAACGG-----GAAGUGGAGUCGAAGGGCGGACACUGCCAUCGAACCGGCUGGAAAGAUGUCAAAAAUGUCUC ...(((((.......(((((.....))))).(((((....)..-----..(((.(..((((..(((((...))))).))))..).))).......)))).....))))). ( -26.50) >consensus GCAAGACGACGCGCAGACAAAAUGUUUGCUAGACAGGAUGUGGCCGGCGAAGUGGGG_CAAAGG___AACUCUGCCACUGGAUGUGGCAGGACGAUGUCAAAAAUGUCUU ..((((((....((((((.....))))))..((((.......((((....(((((...((.((......)).))))))).....)))).......)))).....)))))) (-19.88 = -21.52 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:44 2006