| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 858,621 – 858,735 |
| Length | 114 |
| Max. P | 0.640161 |

| Location | 858,621 – 858,735 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -15.36 |
| Energy contribution | -17.12 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
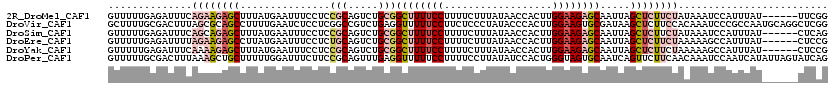
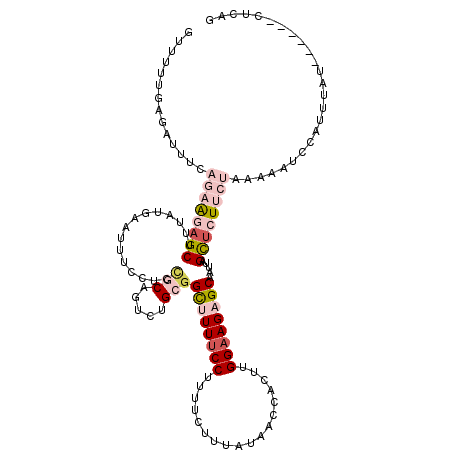
>2R_DroMel_CAF1 858621 114 + 20766785 CCGAA------AUAAAUGGAUUUAUAGAAGAGCUAAUUGCUCUUCCAAGUGGUUAUAAAGAAAAGGAAAAGCCGCAGACUGCGGAGGAAAUUCAUAAAGCUCUUCUGAAAUCUCAAAAAC .....------.....(((((((.((((((((((...((.((((((..((((((...............)))))).(....)))))))....))...))))))))))))))).))..... ( -30.46) >DroVir_CAF1 1270 120 + 1 CCGAGCCUGCAUUGGCGGGAUUUGUGGAAGAGCUUAUCGCACUUCCAAGUGGGUAUAGGGAGAAGGAAAAACCUCAGACGGCCGAGGAGAUUCAAAAGGCUGCGCUAAAGUCGCAAAAGC .(((((((((....))))).))))((((((.((.....)).)))))).((((.(.(((.(.((.((.....))))...(((((...(.....)....)))))).))).).))))...... ( -35.30) >DroSim_CAF1 8846 114 + 1 CUGAG------AUAAAUGGAUUUAUAGAAGAGCUAAUUGCUCUUCCAAGUGGUUAUAAAGAAAAGGAAAAGCCGCAGACUGCGGAGGAAAUUCAUAAAGCUCUGCUGAAAUCUCAAAAAC .((((------((.............(((((((.....)))))))...((((((...............)))))).....((((((.............))))))....))))))..... ( -29.68) >DroEre_CAF1 353 114 + 1 CGGAG------AUAAAUGGCUUUUUAGAAGAGCUAAUUGCUCUUCCAAGUGGUUAUAAAGAAAAGGAAAAGCCGCAGACUGCAGAGGAAAUUCAUAAGGCUCUUCUAAAAUCUCAAAAAC ..(((------((..((((((.(((.(((((((.....))))))).))).))))))........((((.((((((.....)).(((....)))....)))).))))...)))))...... ( -33.80) >DroYak_CAF1 40404 114 + 1 CGGAG------AUAAAUGGCUUUUUAGAAGAGCUAAUUGCUCUUCCAAGUGGUUAUAAAGAAAAGGAAAAGCCGCAGACUGCGGAGGAAAUUCAUAAAGCUCUUUUGAAAUCUCAAAAAC ..(((------((..((((((.(((.(((((((.....))))))).))).)))))).....((((((....((((.....))))..(.....).......))))))...)))))...... ( -29.70) >DroPer_CAF1 373 120 + 1 CUGAUACUAAUAUGAUUGGAUUUGUUGAAGAACUGAUUGCACUACCCAGUGGAUAUAAGGAAAAGGAAAAACCUCAAACUGCGGAAGAAAUCCAAAAAGCAGCUUUAAAGUCGCAAAAAC ...............((((((((.((((....((...(.((((....)))).)....)).....((.....))))))....(....)))))))))...((.(((....))).))...... ( -18.20) >consensus CCGAG______AUAAAUGGAUUUAUAGAAGAGCUAAUUGCUCUUCCAAGUGGUUAUAAAGAAAAGGAAAAGCCGCAGACUGCGGAGGAAAUUCAUAAAGCUCUUCUAAAAUCUCAAAAAC ................(((((((.((((((((((((((((........))))))..........(((....((((.....))))......)))....))))))))))))))).))..... (-15.36 = -17.12 + 1.75)

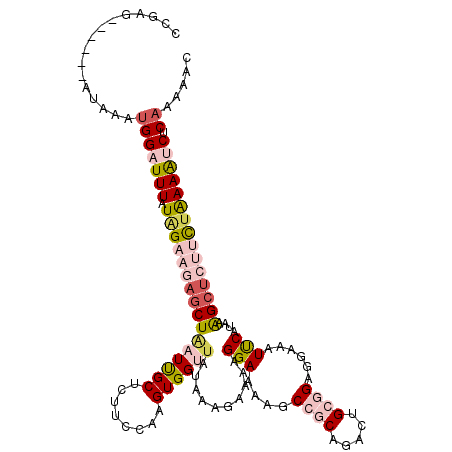
| Location | 858,621 – 858,735 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -14.74 |
| Energy contribution | -16.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 858621 114 - 20766785 GUUUUUGAGAUUUCAGAAGAGCUUUAUGAAUUUCCUCCGCAGUCUGCGGCUUUUCCUUUUCUUUAUAACCACUUGGAAGAGCAAUUAGCUCUUCUAUAAAUCCAUUUAU------UUCGG .....((.(((((.(((((((((..............(((.....)))((((((((..................))))))))....)))))))))..))))))).....------..... ( -30.37) >DroVir_CAF1 1270 120 - 1 GCUUUUGCGACUUUAGCGCAGCCUUUUGAAUCUCCUCGGCCGUCUGAGGUUUUUCCUUCUCCCUAUACCCACUUGGAAGUGCGAUAAGCUCUUCCACAAAUCCCGCCAAUGCAGGCUCGG ((....))........((.(((((...(((...((((((....))))))...)))..................((((((.((.....)).))))))........((....))))))))). ( -30.70) >DroSim_CAF1 8846 114 - 1 GUUUUUGAGAUUUCAGCAGAGCUUUAUGAAUUUCCUCCGCAGUCUGCGGCUUUUCCUUUUCUUUAUAACCACUUGGAAGAGCAAUUAGCUCUUCUAUAAAUCCAUUUAU------CUCAG .....(((((....((.((((((..............(((.....)))((((((((..................))))))))....)))))).)).((((....)))))------)))). ( -26.47) >DroEre_CAF1 353 114 - 1 GUUUUUGAGAUUUUAGAAGAGCCUUAUGAAUUUCCUCUGCAGUCUGCGGCUUUUCCUUUUCUUUAUAACCACUUGGAAGAGCAAUUAGCUCUUCUAAAAAGCCAUUUAU------CUCCG ......((((((((((((((((...............(((.....)))((((((((..................)))))))).....))))))))))).........))------))).. ( -29.87) >DroYak_CAF1 40404 114 - 1 GUUUUUGAGAUUUCAAAAGAGCUUUAUGAAUUUCCUCCGCAGUCUGCGGCUUUUCCUUUUCUUUAUAACCACUUGGAAGAGCAAUUAGCUCUUCUAAAAAGCCAUUUAU------CUCCG ......(((((....(((..(((((...........((((.....)))).......................((((((((((.....)))))))))))))))..)))))------))).. ( -27.90) >DroPer_CAF1 373 120 - 1 GUUUUUGCGACUUUAAAGCUGCUUUUUGGAUUUCUUCCGCAGUUUGAGGUUUUUCCUUUUCCUUAUAUCCACUGGGUAGUGCAAUCAGUUCUUCAACAAAUCCAAUCAUAUUAGUAUCAG ....(((((((((((((.((((.....(((.....))))))))))))))))..............(((((...)))))..)))).................................... ( -19.60) >consensus GUUUUUGAGAUUUCAGAAGAGCUUUAUGAAUUUCCUCCGCAGUCUGCGGCUUUUCCUUUUCUUUAUAACCACUUGGAAGAGCAAUUAGCUCUUCUAAAAAUCCAUUUAU______CUCAG ..............((((((((...............(((.....)))((((((((..................)))))))).....))))))))......................... (-14.74 = -16.55 + 1.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:16 2006