
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,797,332 – 3,797,469 |
| Length | 137 |
| Max. P | 0.999813 |

| Location | 3,797,332 – 3,797,432 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -11.54 |
| Energy contribution | -11.34 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
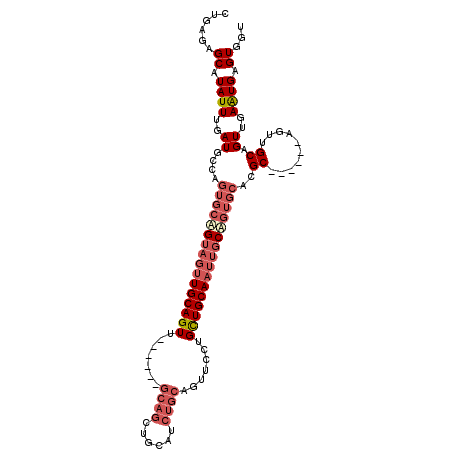
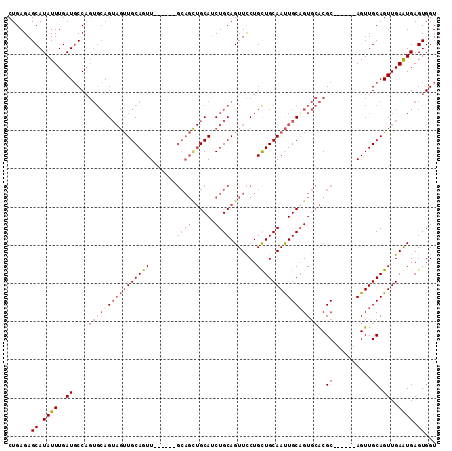
>2R_DroMel_CAF1 3797332 100 + 20766785 CUGGCUUUUGGCGCAUUGUCAGCUGGAGCUUACCACUCAUUCAACUGCAACU------GCGUGCACAGCAAUUGCAACAGGAACUGCAGAUGCAACUGCAGCAGGA ..((((.(..((.........))..)))))..((.....(((...(((((.(------((.......))).)))))....)))((((((......))))))..)). ( -28.10) >DroSec_CAF1 53873 94 + 1 GUGGCUUUUGGCGCAUUGUCAGCUGGAGCUUACCACUCAUUCAACUGCAACU------GCGUGCACUGCAAUUGCAGCAGGAACUGCAGAUGCAGCUGC------A ..........((((((((.(((.((((............)))).)))))).)------))))((((((((.((((((......)))))).))))).)))------. ( -32.90) >DroSim_CAF1 67394 94 + 1 CUGGCUUUUGGCGCAUUGUCAGCUGAAGCUUACCACUCAUUCAACUGCAACU------GCGUGCACUGCAAUUGCAACAGGAACUGCAGAUGCAGCUGC------A ..........((((((((.(((.((((............)))).)))))).)------))))((((((((.(((((........))))).))))).)))------. ( -30.50) >DroEre_CAF1 55221 94 + 1 CUGGCUUUUGGCGCAUUGUCAGCUGGAGCUAACCACUCACUCAACUGCAACUGCAACUGCGCUUACUGCAGUUGCAGCAGAAACUGCAGCUGC------------A .........(((((((((.(((.((.((................)).)).)))))).))))))....((((((((((......))))))))))------------. ( -33.29) >DroWil_CAF1 85052 91 + 1 CCAUCUUUUGUUGCAACAUUGCACAAGGCUUAUCAUUCAUGCAAGUGCAAUUGCAAUUG---------GAAUUGCAAUCGCAACUGCUGAUGCUGUUGC------A ...(((...((((((..(((((((...((...........))..))))))))))))).)---------)).........(((((.((....)).)))))------. ( -27.90) >consensus CUGGCUUUUGGCGCAUUGUCAGCUGGAGCUUACCACUCAUUCAACUGCAACU______GCGUGCACUGCAAUUGCAACAGGAACUGCAGAUGCAGCUGC______A ..((((((.(((.........))))))))).....................................(((.(((((........))))).)))............. (-11.54 = -11.34 + -0.20)

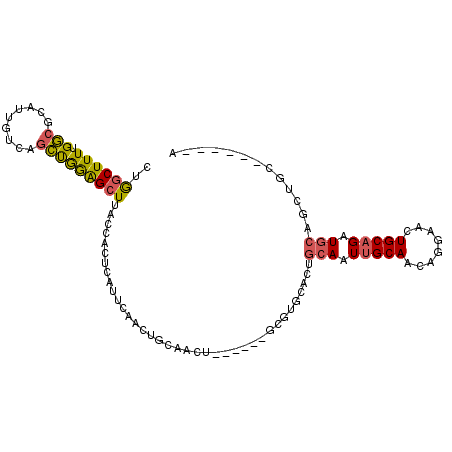
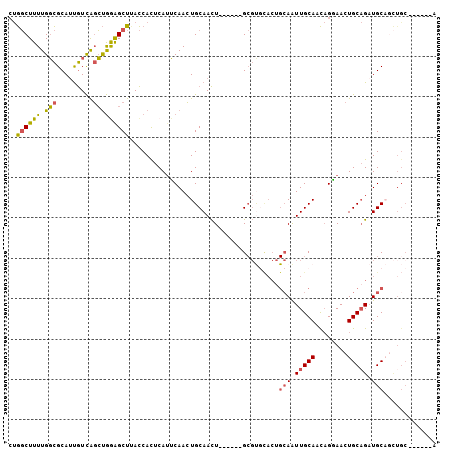
| Location | 3,797,332 – 3,797,432 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Mean single sequence MFE | -35.76 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.43 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3797332 100 - 20766785 UCCUGCUGCAGUUGCAUCUGCAGUUCCUGUUGCAAUUGCUGUGCACGC------AGUUGCAGUUGAAUGAGUGGUAAGCUCCAGCUGACAAUGCGCCAAAAGCCAG ...(((.(((((((((...((((...)))))))))))))...)))(((------(.(((((((((...((((.....)))))))))).)))))))........... ( -34.20) >DroSec_CAF1 53873 94 - 1 U------GCAGCUGCAUCUGCAGUUCCUGCUGCAAUUGCAGUGCACGC------AGUUGCAGUUGAAUGAGUGGUAAGCUCCAGCUGACAAUGCGCCAAAAGCCAC (------(((.(((((..((((((....))))))..)))))))))(((------(.(((((((((...((((.....)))))))))).)))))))........... ( -38.70) >DroSim_CAF1 67394 94 - 1 U------GCAGCUGCAUCUGCAGUUCCUGUUGCAAUUGCAGUGCACGC------AGUUGCAGUUGAAUGAGUGGUAAGCUUCAGCUGACAAUGCGCCAAAAGCCAG (------(((.(((((..(((((......)))))..)))))))))(((------(.((((((((((...(((.....)))))))))).)))))))........... ( -33.50) >DroEre_CAF1 55221 94 - 1 U------------GCAGCUGCAGUUUCUGCUGCAACUGCAGUAAGCGCAGUUGCAGUUGCAGUUGAGUGAGUGGUUAGCUCCAGCUGACAAUGCGCCAAAAGCCAG .------------.((((((((......(((((((((((.......))))))))))))))))))).(((....((((((....))))))....))).......... ( -40.20) >DroWil_CAF1 85052 91 - 1 U------GCAACAGCAUCAGCAGUUGCGAUUGCAAUUC---------CAAUUGCAAUUGCACUUGCAUGAAUGAUAAGCCUUGUGCAAUGUUGCAACAAAAGAUGG (------((((((......(((((((((((((......---------)))))))))))))..(((((..(..........)..))))))))))))........... ( -32.20) >consensus U______GCAGCUGCAUCUGCAGUUCCUGUUGCAAUUGCAGUGCACGC______AGUUGCAGUUGAAUGAGUGGUAAGCUCCAGCUGACAAUGCGCCAAAAGCCAG .......((....(((..((((((....))))))..)))................((((((((((...((((.....)))))))))).))))))............ (-15.48 = -15.92 + 0.44)

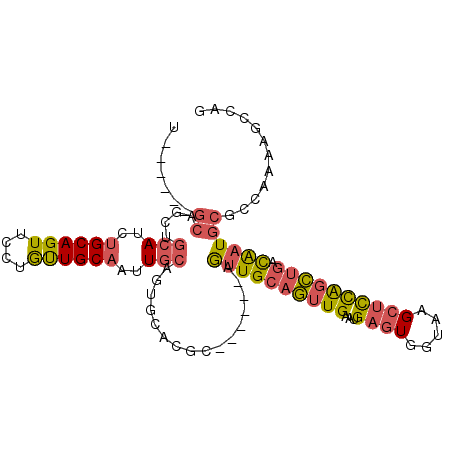
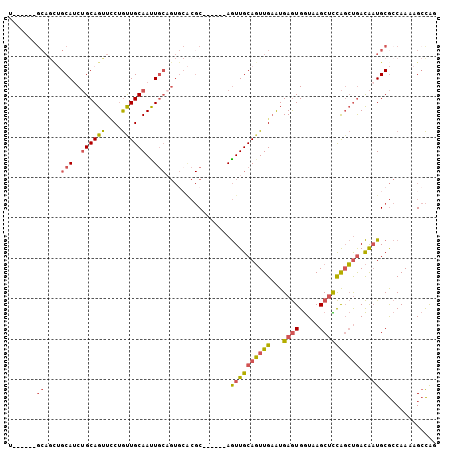
| Location | 3,797,363 – 3,797,469 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -19.95 |
| Energy contribution | -22.89 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3797363 106 - 20766785 CUGAGAGCAUAUUUGAUGCCAGUGCAGUAGUUGCAGUUCCUGCUGCAGUUGCAUCUGCAGUUCCUGUUGCAAUUGCUGUGCACGC------AGUUGCAGUUGAAUGAGUGGU ......((.((((..((....(..((((((((((((.....(((((((......))))))).....))))))))))))..)..((------....)).))..)))).))... ( -39.50) >DroSec_CAF1 53904 100 - 1 CUGAGAGCAUAUUUGAUUCCAGUGCAGUAGUUGCAGUU------GCAGCUGCAUCUGCAGUUCCUGCUGCAAUUGCAGUGCACGC------AGUUGCAGUUGAAUGAGUGGU ......((.((((..(((...((((((((((((((((.------(.((((((....)))))).).)))))))))))..)))))((------....)))))..)))).))... ( -38.40) >DroSim_CAF1 67425 100 - 1 CUGAGAGCAUAUUUGAUGCCAGUGCAGUAGUUGCAGUU------GCAGCUGCAUCUGCAGUUCCUGUUGCAAUUGCAGUGCACGC------AGUUGCAGUUGAAUGAGUGGU ......((.((((..((....(((((((((((((....------))))))))..((((((((.(....).)))))))))))))((------....)).))..)))).))... ( -35.20) >DroEre_CAF1 55252 92 - 1 CUGAGAGCAUAUUUGAUGCC-----UG---CUGCAGUU------------GCAGCUGCAGUUUCUGCUGCAACUGCAGUAAGCGCAGUUGCAGUUGCAGUUGAGUGAGUGGU ...((.((((.....)))))-----)(---(((((..(------------((((((((.(((..((((((....))))))))))))))))))..))))))............ ( -39.40) >consensus CUGAGAGCAUAUUUGAUGCCAGUGCAGUAGUUGCAGUU______GCAGCUGCAUCUGCAGUUCCUGCUGCAAUUGCAGUGCACGC______AGUUGCAGUUGAAUGAGUGGU ......((.((((..((....((((((((((((((((.......((((......)))).......))))))))))))))))..((..........)).))..)))).))... (-19.95 = -22.89 + 2.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:39 2006