| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,778,331 – 3,778,435 |
| Length | 104 |
| Max. P | 0.767551 |

| Location | 3,778,331 – 3,778,435 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -24.95 |
| Energy contribution | -26.48 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
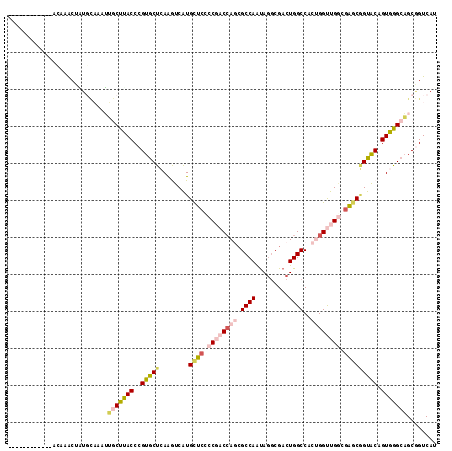
>2R_DroMel_CAF1 3778331 104 + 20766785 ------------ACAAACUAUGCAUAUUGCUUACCCGUGCUCAAGUCAUGCUCCCCGACCAGCGCCAAUAGGCGACUGGCCACUGGUUGGCGAGCGGUACAGUGGGCAGCGGUCAU ------------.........((.....))....(((((((((.....(((((.((((((((.((((...(....)))))..)))))))).)))))......)))))).))).... ( -40.40) >DroSec_CAF1 34471 104 + 1 ------------AUAAACUGUGCAAAUUGCUUACCCGUGCUCAAGUCAUGCUCCCCGACCAACGCCAAUAGGCGACUGGCCACUGGUUGGCGAGCGGUACAGUGGGCAGCGGUCAU ------------.........((.....))....(((((((((.....(((((.(((((((..((((...(....)))))...))))))).)))))......)))))).))).... ( -37.50) >DroSim_CAF1 56945 104 + 1 ------------ACAAACUAUGCAUAUUGCUUACCCGUGCUCAAGUCAUGCUGACCGACCAGCGCCAAUAGGCGACUGGCCACUGGUUGGUGAGCGGUACAGUGGGCAGCGGUCAU ------------.........((.....))....(((((((((.....((((.(((((((((.((((...(....)))))..))))))))).))))......)))))).))).... ( -41.10) >DroEre_CAF1 36299 116 + 1 AUUCCAAAUACAACACACUGUGCAAAUUGCUCACCCGUGCUCAAGUCAUGCUCCCCGACCAGCGCCAAUAGGCGACUGGCCACUGGUUGGCGAGCGGUACAGUGGGCAGCGGUCAU ...............((((((((...(((((.(((.(((..((.(((..(((........)))(((....))))))))..))).))).)))))...))))))))(((....))).. ( -42.10) >DroAna_CAF1 33158 111 + 1 --UCCUAUUAU--UAAGGUCU-UUAAGGACUUACCCGUACUGAGAUCAUGUUCUCCCACCAAGGCCAAUAAGCGACUGGCCACCGGCUGCCGGGCAGUACAGUGAGCUGCAGUCAU --.........--...(((((-(...((......)).....((((......)))).....)))))).......((((((((((..(((((...)))))...))).))..))))).. ( -29.90) >DroPer_CAF1 27815 95 + 1 ------------AUGGAC---------CACUCACCCGUGCUGAGAUCGUGCUCUCCCACGAGCGCCAGCAGACGACUGGCCACCGGCUGACGGACCGUGCAGUGGGCAGCUGUCAC ------------((((..---------.......))))((((.....((((((......)))))))))).((((.(((.((((((((.....).)))....)))).))).)))).. ( -33.10) >consensus ____________ACAAACUAUGCAAAUUGCUUACCCGUGCUCAAGUCAUGCUCCCCGACCAGCGCCAAUAGGCGACUGGCCACUGGUUGGCGAGCGGUACAGUGGGCAGCGGUCAU ...........................(((((((..(((((........((((.((((((((.((((.........))))..)))))))).))))))))).)))))))........ (-24.95 = -26.48 + 1.53)


| Location | 3,778,331 – 3,778,435 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
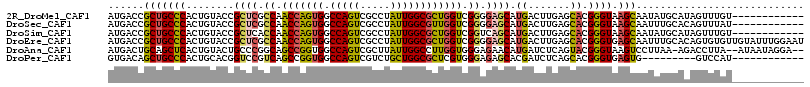
>2R_DroMel_CAF1 3778331 104 - 20766785 AUGACCGCUGCCCACUGUACCGCUCGCCAACCAGUGGCCAGUCGCCUAUUGGCGCUGGUCGGGGAGCAUGACUUGAGCACGGGUAAGCAAUAUGCAUAGUUUGU------------ ....(((.(((.((..((...((((.((.(((((((.(((((.....)))))))))))).)).))))...)).)).))))))....((.....)).........------------ ( -40.00) >DroSec_CAF1 34471 104 - 1 AUGACCGCUGCCCACUGUACCGCUCGCCAACCAGUGGCCAGUCGCCUAUUGGCGUUGGUCGGGGAGCAUGACUUGAGCACGGGUAAGCAAUUUGCACAGUUUAU------------ ....(((.(((.((..((...((((.((.(....)((((((.((((....)))))))))))).))))...)).)).))))))((((((..........))))))------------ ( -35.90) >DroSim_CAF1 56945 104 - 1 AUGACCGCUGCCCACUGUACCGCUCACCAACCAGUGGCCAGUCGCCUAUUGGCGCUGGUCGGUCAGCAUGACUUGAGCACGGGUAAGCAAUAUGCAUAGUUUGU------------ ....(((.(((.((..((...(((.(((.(((((((.(((((.....)))))))))))).))).)))...)).)).))))))....((.....)).........------------ ( -36.40) >DroEre_CAF1 36299 116 - 1 AUGACCGCUGCCCACUGUACCGCUCGCCAACCAGUGGCCAGUCGCCUAUUGGCGCUGGUCGGGGAGCAUGACUUGAGCACGGGUGAGCAAUUUGCACAGUGUGUUGUAUUUGGAAU (..(((((((.....((((..(((((((..((..(((((((.((((....))))))))))).)).((.........))...)))))))....))))))))).))..)......... ( -43.80) >DroAna_CAF1 33158 111 - 1 AUGACUGCAGCUCACUGUACUGCCCGGCAGCCGGUGGCCAGUCGCUUAUUGGCCUUGGUGGGAGAACAUGAUCUCAGUACGGGUAAGUCCUUAA-AGACCUUA--AUAAUAGGA-- ..((((((((....))))..((((((((.(((((.(((((((.....))))))))))))..((((......)))).)).)))))))))).....-...(((..--.....))).-- ( -36.80) >DroPer_CAF1 27815 95 - 1 GUGACAGCUGCCCACUGCACGGUCCGUCAGCCGGUGGCCAGUCGUCUGCUGGCGCUCGUGGGAGAGCACGAUCUCAGCACGGGUGAGUG---------GUCCAU------------ (((.(((.......))))))((.(((((.(((((..(.(....).)..)))))(((((((.((((......))))..))))))))).))---------).))..------------ ( -39.90) >consensus AUGACCGCUGCCCACUGUACCGCUCGCCAACCAGUGGCCAGUCGCCUAUUGGCGCUGGUCGGGGAGCAUGACUUGAGCACGGGUAAGCAAUUUGCACAGUUUAU____________ ......(((((((........((((.((.(((((((.(((((.....)))))))))))).)).)))).((.......)).)))).)))............................ (-27.94 = -27.78 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:34 2006