| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,770,187 – 3,770,278 |
| Length | 91 |
| Max. P | 0.698406 |

| Location | 3,770,187 – 3,770,278 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -12.31 |
| Energy contribution | -13.31 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
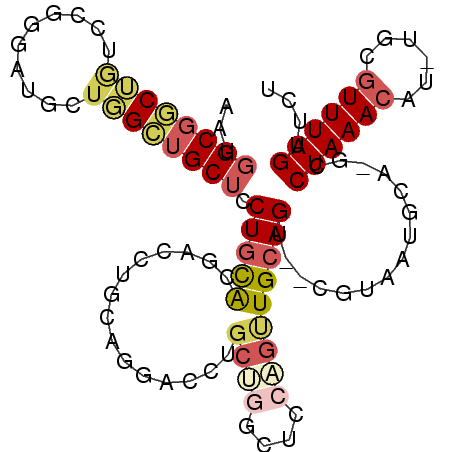
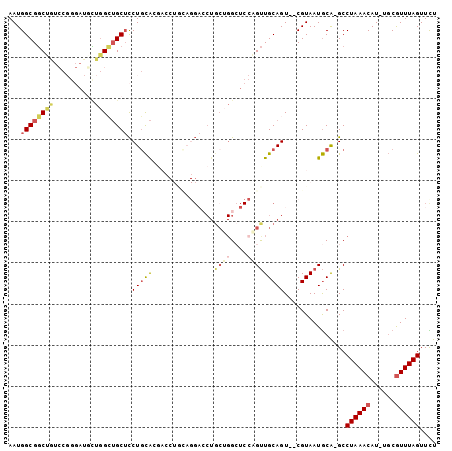
>2R_DroMel_CAF1 3770187 91 + 20766785 AAUGGCGGCUAUCCGGGAUGUUGGGUGCUCCUGCAG---------GACCUGCUGGCUCCAGUUGCAGU--CGUAAUGCA-GCCUAAACAU-UGCGUUUAGUUCU ......(((((..((.((((((.((.(((...((((---------...)))).))).)).((((((..--.....))))-))...)))))-).))..))))).. ( -30.30) >DroVir_CAF1 27728 81 + 1 AAUGGCGUCGA-------UGAUGGGGGCAUCUCUGCGA----------CUGCGGG----CGCUGCAGU--CGUAAUACGCGCCUAAACAUUGCCAUUUAGUUUU ((((((...((-------((.((((.((.....(((((----------((((((.----..)))))))--))))....)).))))..))))))))))....... ( -31.80) >DroSec_CAF1 26164 100 + 1 AAUGGCGGCUGUCCGGGAUGCUGGCUGCUCCUGCAGGACCUGCAGGACCUGCUGGCUCCAGUUGCAGU--CGUAAUGCA-GCCUAAACAU-UGCGUUUAGUUCU ....((((((((...(((.((..((.(.((((((((...)))))))).).))..)))))....)))))--)))......-..((((((..-...)))))).... ( -43.40) >DroSim_CAF1 48234 100 + 1 AAUGGCGGCUGUCCGGGAUGCUGGCUGCUCCUGCAGGACCUGCAGGACCUGCUGGCUCCAGUUGCAGU--CGUAAUGCA-GCCUAAACAU-UGCGUUUAGUUCU ....((((((((...(((.((..((.(.((((((((...)))))))).).))..)))))....)))))--)))......-..((((((..-...)))))).... ( -43.40) >DroYak_CAF1 20230 100 + 1 AAUGGCGGCUGUCCGGGAUCCUGGCUGCUCCUGCACGAGCUACAGGACCUGCUGGCUCCUGUUGCAGU--CGUAAUGCA-GCCUAAACAU-UGCGUUUAGUUCU ......(((((..((.(((...((((((...(((((..((.((((((((....)).)))))).)).))--.)))..)))-))).....))-).))..))))).. ( -36.20) >DroAna_CAF1 26139 89 + 1 AAUGGCGGCGCUGC-----UUUUGCUGCUCCUGCACUUCCUGUAGG--------GGUCCCGUUGCAGUCCCGUAAUGCA-GCCUAAACAU-GGCGUUUAGUUUU ....((((.(((((-----...((..(((((((((.....))))))--------)))..))..))))).))))......-(((.......-))).......... ( -32.40) >consensus AAUGGCGGCUGUCCGGGAUGCUGGCUGCUCCUGCACGACCUGCAGGACCUGCUGGCUCCAGUUGCAGU__CGUAAUGCA_GCCUAAACAU_UGCGUUUAGUUCU ...((((((((..........)))))))).(((((...............((((....)))))))))...............((((((......)))))).... (-12.31 = -13.31 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:30 2006