
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,755,826 – 3,755,958 |
| Length | 132 |
| Max. P | 0.990190 |

| Location | 3,755,826 – 3,755,929 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.91 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -18.48 |
| Energy contribution | -21.07 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
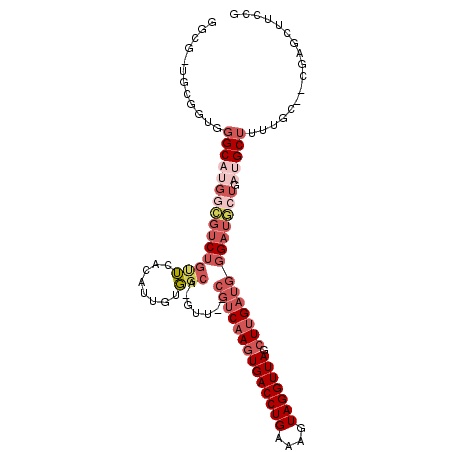
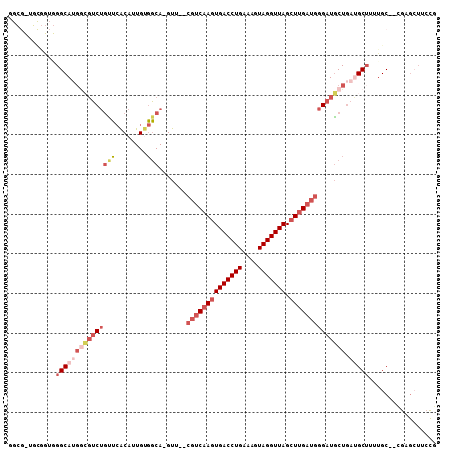
>2R_DroMel_CAF1 3755826 103 + 20766785 GGCGUUGCGGUGGGCAUGGCGUCUGUUCACAUUGUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC--CGAGCUUCCG ((.(((.(((((((((((((((((((.(((...))))).-...--((((((((((((((....))))))).)))))))))))))).))))))..))--)))))..)). ( -40.30) >DroSec_CAF1 12527 87 + 1 ------------GGCA----GUCUGUUCACAUUGUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGCGAUGCUGAUGCUUUUGC--CGAGCUUCCG ------------.(((----((........)))))((.(-(((--((((((((((((((....))))))).))))))((((.((....))..))))--.))))).)). ( -28.70) >DroSim_CAF1 6778 91 + 1 ------------GGCAUGGCGUCUGUUCACAUUGUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC--CGAGCUUCCG ------------((((.((((((....(((...)))(((-.((--((((((((((((((....))))))).))))))))).))).))))))..)))--)......... ( -36.40) >DroEre_CAF1 12368 93 + 1 GGCGUUGCGGUGGGCAUGGCGUCUGCUCACAUUGUGGCA-GUU--CGUCAAGUGACCUGAAUGUAGGUUAGCUUGAUGGGAUGCUGAUGCUGAUGC------------ .(((((((((((((((.......))))))).....((((-.((--((((((((((((((....))))))).))))))))).))))..))).)))))------------ ( -38.00) >DroYak_CAF1 5985 105 + 1 GGCGCUGCGGUGGGCAUGGCGUCUGCUCACAUUGAGGCA-GUU--CGUCAAGUGACCUGAAUGUAGGUUAGCUUGAUGGGAUGCUGAAGCUUUUGCUCUGUGCUUCCG (((((.((((((((((.......))))))).....((((-.((--((((((((((((((....))))))).))))))))).))))........)))...))))).... ( -40.60) >DroAna_CAF1 12019 99 + 1 UGGGCUUUGGGGGGCAGGAACG--AAGAGCAAUGCCCCAUGUUUGUAUCAAGUGACCUGAAUGUAGGUUAGGUGG-----AUUCUGAGGCAGACGC--AGAGAUAGUG ....(((((.((((((....(.--....)...))))))..((((((.(((..(.((((((.......)))))).)-----....))).)))))).)--))))...... ( -30.30) >consensus GGCG_UGCGGUGGGCAUGGCGUCUGUUCACAUUGUGGCA_GUU__CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC__CGAGCUUCCG ............(((((((((((((((........))).......((((((((((((((....))))))).)))))))))))))).)))))................. (-18.48 = -21.07 + 2.59)

| Location | 3,755,859 – 3,755,958 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.71 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
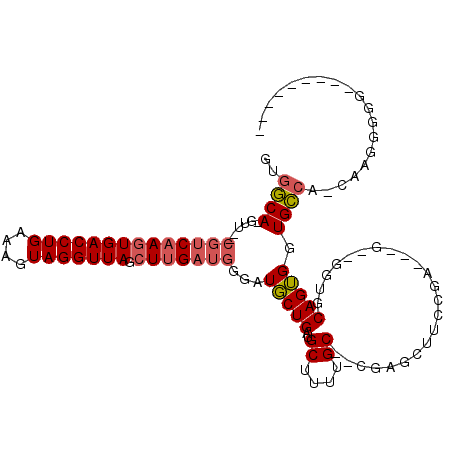
>2R_DroMel_CAF1 3755859 99 + 20766785 GUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC--CGAGCUUCCGAGGAG--GGUGCAGUGGUGCCCCCAAGGGGG--------- .(((((-(((--((((((((((((((....))))))).)))))))))..((....))..))))--))..(((((..((.(--((..(....)..)))))..)))))--------- ( -42.20) >DroSec_CAF1 12544 96 + 1 GUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGCGAUGCUGAUGCUUUUGC--CGAGCUUCCGA---G--GGUGCAGUGGUGCCCCCAAGGGGG--------- .(((((-(..--((((((((((((((....))))))).)))))))....((....))..))))--))..(((((..---(--((.(((....))).)))..)))))--------- ( -36.30) >DroSim_CAF1 6799 96 + 1 GUGGCA-GUU--CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC--CGAGCUUCCGA---G--GGUGCAGUGGUGCCCCCAAGGGGG--------- .(((((-(((--((((((((((((((....))))))).)))))))))..((....))..))))--))..(((((..---(--((.(((....))).)))..)))))--------- ( -38.30) >DroEre_CAF1 12401 92 + 1 GUGGCA-GUU--CGUCAAGUGACCUGAAUGUAGGUUAGCUUGAUGGGAUGCUGAUGCUGAUGC-------------------AACGCAGUGGUGCCA-CAAGGGGGUUUAAUGUU ((((((-.((--((((((((((((((....))))))).)))))))))..((((.(((....))-------------------)...))))..)))))-)................ ( -34.20) >DroYak_CAF1 6018 106 + 1 GAGGCA-GUU--CGUCAAGUGACCUGAAUGUAGGUUAGCUUGAUGGGAUGCUGAAGCUUUUGCUCUGUGCUUCCGA---G--AGUGCAGUGGUGCCA-CAAGGGGGCUUGAGGUU ..((((-.((--((((((((((((((....))))))).))))))))).))))..((((((.((((((..(((....---)--))..).(((....))-)...)))))..)))))) ( -40.80) >DroAna_CAF1 12050 104 + 1 GCCCCAUGUUUGUAUCAAGUGACCUGAAUGUAGGUUAGGUGG-----AUUCUGAGGCAGACGC--AGAGAUAGUGCUGAUGUCUGGCAGGGGUGGCA-GGAGAGGGAU-AAUG-- ..(((...(((((.(((..(((((((....)))))))..)))-----(((((..(.(((((((--((........))).)))))).).))))).)))-))...)))..-....-- ( -32.30) >consensus GUGGCA_GUU__CGUCAAGUGACCUGAAAGUAGGUUAGCUUGAUGGGAUGCUGAUGCUUUUGC__CGAGCUUCCGA___G__GGUGCAGUGGUGCCA_CAAGGGGG_________ ..((((......((((((((((((((....))))))).)))))))...(((((..((....)).......................))))).))))................... (-18.26 = -18.71 + 0.45)


| Location | 3,755,859 – 3,755,958 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
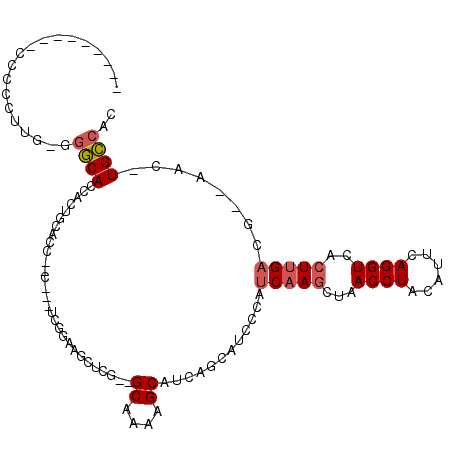
>2R_DroMel_CAF1 3755859 99 - 20766785 ---------CCCCCUUGGGGGCACCACUGCACC--CUCCUCGGAAGCUCG--GCAAAAGCAUCAGCAUCCCAUCAAGCUAACCUACUUUCAGGUCACUUGACG--AAC-UGCCAC ---------((((....)))).......(((..--......(((.(((..--((....))...))).)))..(((((...((((......))))..)))))..--...-)))... ( -25.30) >DroSec_CAF1 12544 96 - 1 ---------CCCCCUUGGGGGCACCACUGCACC--C---UCGGAAGCUCG--GCAAAAGCAUCAGCAUCGCAUCAAGCUAACCUACUUUCAGGUCACUUGACG--AAC-UGCCAC ---------...((..(((.(((....))).))--)---..))......(--(((...((....)).(((..(((((...((((......))))..)))))))--)..-)))).. ( -27.40) >DroSim_CAF1 6799 96 - 1 ---------CCCCCUUGGGGGCACCACUGCACC--C---UCGGAAGCUCG--GCAAAAGCAUCAGCAUCCCAUCAAGCUAACCUACUUUCAGGUCACUUGACG--AAC-UGCCAC ---------((((....)))).......(((..--.---(((((.(((..--((....))...))).)))..(((((...((((......))))..))))).)--)..-)))... ( -25.90) >DroEre_CAF1 12401 92 - 1 AACAUUAAACCCCCUUG-UGGCACCACUGCGUU-------------------GCAUCAGCAUCAGCAUCCCAUCAAGCUAACCUACAUUCAGGUCACUUGACG--AAC-UGCCAC ................(-(((((....(((..(-------------------((....)))...))).....(((((...((((......))))..)))))..--...-)))))) ( -21.80) >DroYak_CAF1 6018 106 - 1 AACCUCAAGCCCCCUUG-UGGCACCACUGCACU--C---UCGGAAGCACAGAGCAAAAGCUUCAGCAUCCCAUCAAGCUAACCUACAUUCAGGUCACUUGACG--AAC-UGCCUC ..((.((((....))))-.)).......(((..--.---(((((.((...((((....))))..)).)))..(((((...((((......))))..))))).)--)..-)))... ( -23.20) >DroAna_CAF1 12050 104 - 1 --CAUU-AUCCCUCUCC-UGCCACCCCUGCCAGACAUCAGCACUAUCUCU--GCGUCUGCCUCAGAAU-----CCACCUAACCUACAUUCAGGUCACUUGAUACAAACAUGGGGC --....-..........-.(((.((.(((.(((((..(((........))--).)))))...))).((-----(......((((......)))).....)))........))))) ( -17.00) >consensus _________CCCCCUUG_GGGCACCACUGCACC__C___UCGGAAGCUCG__GCAAAAGCAUCAGCAUCCCAUCAAGCUAACCUACAUUCAGGUCACUUGACG__AAC_UGCCAC ...................((((.............................((....))............(((((...((((......))))..)))))........)))).. (-11.04 = -11.43 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:25 2006