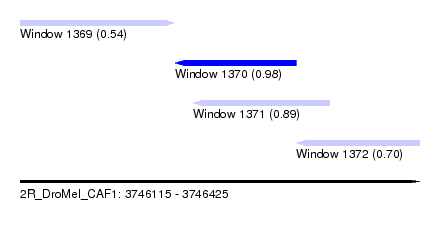
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,746,115 – 3,746,425 |
| Length | 310 |
| Max. P | 0.980357 |

| Location | 3,746,115 – 3,746,235 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -30.79 |
| Energy contribution | -30.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3746115 120 + 20766785 UGCUGCUGUGGCUUCUCCUCUUUCUUUCUUUCCUUUUUUUUCUUAUUCCCGGCCCAGGGAUCAAGGUAAACGCCAUUUAGUGUGGCACGAUCGCUGACGAUCGGUAGCCAGGCUUGUAUC (((.(((.(((((...((......................((((..((((......))))..)))).....(((((.....)))))..(((((....))))))).))))))))..))).. ( -35.50) >DroSim_CAF1 8917 119 + 1 UGCUGCUGUGGCUUCUCCUCUUUCUUUCCUUCCUUUUUUCU-UUAUUCCCGGCCCAGGGAUCAAGGUAAACGCCAUUUAGUGUGGCACGAUCGCUGCCGAUCGGUAGCCAGGCUUGUAUC (((.(((.(((((...((.................(((.((-((..((((......))))..)))).))).(((((.....)))))..(((((....))))))).))))))))..))).. ( -34.80) >DroYak_CAF1 8653 112 + 1 U---GCUGUGGCUUCUCUUCGCUCUUU----CCUUUUUUCU-UCAUUCCCGGCUCAGGGAUCAAGGUAAACGCCAUUAAGUGUGGCACGAUCGCUGCCGAUCGGUAGCCAGGCCUGUAUC (---((...(((((....(((......----.......(((-(...((((......))))..)))).....(((((.....))))).)))..((((((....)))))).))))).))).. ( -33.20) >consensus UGCUGCUGUGGCUUCUCCUCUUUCUUUC_UUCCUUUUUUCU_UUAUUCCCGGCCCAGGGAUCAAGGUAAACGCCAUUUAGUGUGGCACGAUCGCUGCCGAUCGGUAGCCAGGCUUGUAUC ....(((.(((((.(................((((...........((((......))))..)))).....(((((.....))))).((((((....))))))).))))))))....... (-30.79 = -30.79 + -0.00)

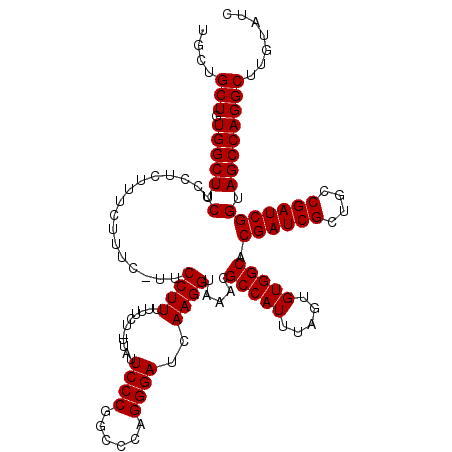
| Location | 3,746,235 – 3,746,329 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -26.05 |
| Energy contribution | -25.83 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3746235 94 - 20766785 AAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGAAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAGGA------------------------CUG--ACAGAUACA .......((((((...........((((((((((....))))))).)))((.(((...((.(((....))).....)).)))------------------------)).--..)))))). ( -28.10) >DroSim_CAF1 9036 118 - 1 AAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAGGACCGGCAGACACAGUACACAUCGUACUG--ACAGAUACA .......((((((....((((...((((((((((....))))))).)))........(((.((....((((.....)).)).)))))..))))(((((.....))))).--..)))))). ( -38.40) >DroYak_CAF1 8765 111 - 1 AAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCCAGCCCGGGAAGCCCCAAAAAGGAGGACUGGCAGAC---------ACAGUACAGAUACAAAUACA .......((((((...(((((...((((((((((....))))))).))).....((((.((((....))((.....)).)).))))...))---------)))....))))))....... ( -35.60) >consensus AAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAGGAC_GGCAGAC_________A__GUACUG__ACAGAUACA .......((((((...........((((((((((....))))))).)))...(((...((.(((....))).....)).)))...............................)))))). (-26.05 = -25.83 + -0.22)

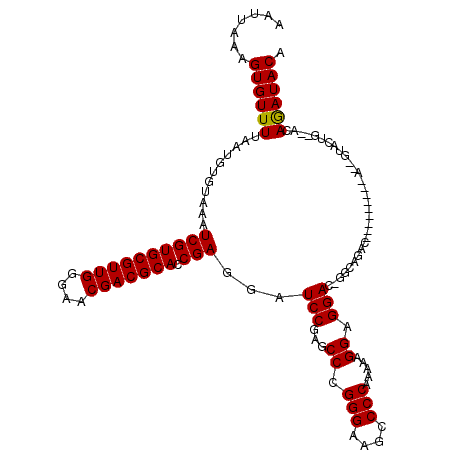
| Location | 3,746,249 – 3,746,355 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3746249 106 - 20766785 UCCCAA--------------CCGACCGGGUGUACAAUAAAAAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGAAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAG (((...--------------......((((.((((...(((((......)))))....))))..((((((((((....))))))).)))........))))(((....))).....))). ( -32.90) >DroSim_CAF1 9074 106 - 1 UCCCAG--------------CCGACCGGGUGUACAAUAAAAAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAG .....(--------------(...((((((.((((...(((((......)))))....))))..((((((((((....))))))).)))........))))))...)).((.....)).. ( -32.30) >DroYak_CAF1 8796 120 - 1 UCCCAGCUUCGUAUGUACACCCGACCGGGUGUACAAUAAAAAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCCAGCCCGGGAAGCCCCAAAAAGGAG (((...(((((..(((((((((....)))))))))...............................((((((((....))))))))))))).((((.....))))...........))). ( -43.10) >consensus UCCCAG______________CCGACCGGGUGUACAAUAAAAAUUAAAGUGUUUUAAUGUGUAAAUCGUGCGUUGGGAACGACGCACCGAGGAUCCGAGCCCGGGAAGCCCCAAAAAGGAG (((.......................((((.((((...(((((......)))))....))))..((((((((((....))))))).)))........))))(((....))).....))). (-31.90 = -31.90 + 0.00)

| Location | 3,746,329 – 3,746,425 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3746329 96 - 20766785 AGUCUGCCGGUGC----------CCUCAGAAUUGAUUAAAAAAGAGGGAAGUAGAUCGUCACUGGUCGACAGAUCGAAGCUCCCAA--------------CCGACCGGGUGUACAAUAAA .(((.((((((((----------((((................)))))...........))))))).)))........((.(((..--------------......))).))........ ( -25.39) >DroSim_CAF1 9154 96 - 1 AGUCCGCCGGUGC----------CCUCAGAAUUGAUUAAAAAAGAGGGAAGUAGAUCGUCACUGGUCGACAGAUCGAAGCUCCCAG--------------CCGACCGGGUGUACAAUAAA .(((.((((((((----------((((................)))))...........))))))).)))........((.(((..--------------......))).))........ ( -25.39) >DroYak_CAF1 8876 120 - 1 AGUCCGCCGGUGUCUUUACUCUGCCUCAGAAUUGAUUAAAAAAGAGGGAAGUAGAUCGUCACUGGUCGACAGAUCGAAGCUCCCAGCUUCGUAUGUACACCCGACCGGGUGUACAAUAAA .(((.(((((((.(((((((.(.((((................)))).)))))))..).))))))).)))....((((((.....))))))..(((((((((....)))))))))..... ( -47.99) >consensus AGUCCGCCGGUGC__________CCUCAGAAUUGAUUAAAAAAGAGGGAAGUAGAUCGUCACUGGUCGACAGAUCGAAGCUCCCAG______________CCGACCGGGUGUACAAUAAA .((.(((((((..............((......))..........((((.((.(((((((.......))).))))...))))))...................))).)))).))...... (-22.85 = -22.63 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:14 2006