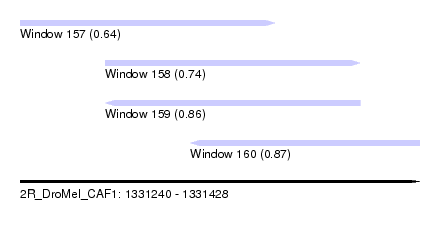
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,331,240 – 1,331,428 |
| Length | 188 |
| Max. P | 0.871958 |

| Location | 1,331,240 – 1,331,360 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1331240 120 + 20766785 CCUUUCCUGCAGCAGCACAUGAAAAUGUUUAAAUAUUUAUGAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUA ......((..((((...((((((.((......)).))))))......))))..)).......((((((...(((......)))...))))))..((((((((.((.....)))))))))) ( -22.10) >DroSec_CAF1 49767 120 + 1 CCUUUCCUGCAGCAGCACAUGAAAAUGUUUAAAUAUUUAUGAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUA ......((..((((...((((((.((......)).))))))......))))..)).......((((((...(((......)))...))))))..((((((((.((.....)))))))))) ( -22.10) >DroEre_CAF1 31423 120 + 1 CCUUUUCUGUAGCAGCACAUGAAAAUGUUUAAAUAUUUAUGAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUA ......((.(((((...((((((.((......)).))))))......))))).)).......((((((...(((......)))...))))))..((((((((.((.....)))))))))) ( -23.20) >DroYak_CAF1 16741 120 + 1 CCUUUUUUGUAGCAGCACAUGAAAAUAUUUAAAUAUUUAUGAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUA ..............(((.((((((((((((.........(((((((.......)))))))..((((((...(((......)))...))))))........)))))).)))))).)))... ( -23.00) >consensus CCUUUCCUGCAGCAGCACAUGAAAAUGUUUAAAUAUUUAUGAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUA ..............(((.((((((((((((.........(((((((.......)))))))..((((((...(((......)))...))))))........)))))).)))))).)))... (-22.44 = -22.25 + -0.19)

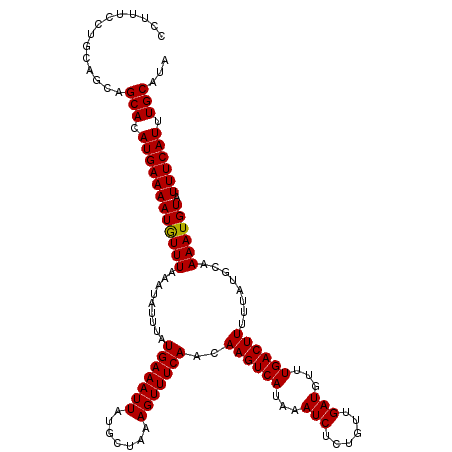
| Location | 1,331,280 – 1,331,400 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -25.19 |
| Energy contribution | -25.19 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1331280 120 + 20766785 GAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUAAAAAUGGCUGACAAAAUAUUUUCCAAGCCGAUAUUCAUGG ((((((.......))))))...((((((...(((......)))...))))))((((((((((.((.....))))))))))))..(((((................))))).......... ( -25.19) >DroSec_CAF1 49807 120 + 1 GAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUAAAAAUGGCUGACAAAAUAUUUUCCAAGCCGAUAUUCAUGG ((((((.......))))))...((((((...(((......)))...))))))((((((((((.((.....))))))))))))..(((((................))))).......... ( -25.19) >DroEre_CAF1 31463 120 + 1 GAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUAAAAAUGGCUGACAAAAUAUUAUCCAAGCCGAUGUUCAGGG ((((((.......))))))((((.((((...(((......)))...))))((((((((((((.((.....))))))))))))))(((((................))))).))))..... ( -25.89) >DroYak_CAF1 16781 120 + 1 GAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUAAAAAUGGCUGACAAAAUAUUUUCCAAGCCGAUAUUCAUGG ((((((.......))))))...((((((...(((......)))...))))))((((((((((.((.....))))))))))))..(((((................))))).......... ( -25.19) >consensus GAAAUUAUGCUAAAGUUUCAACAAGUCAUAAAUCUCUGUUGAUGUUUGACUUUUUAUGCAAAAUGUAUUUCAUUUGCAUAAAAAUGGCUGACAAAAUAUUUUCCAAGCCGAUAUUCAUGG ((((((.......))))))...((((((...(((......)))...))))))((((((((((.((.....))))))))))))..(((((................))))).......... (-25.19 = -25.19 + -0.00)

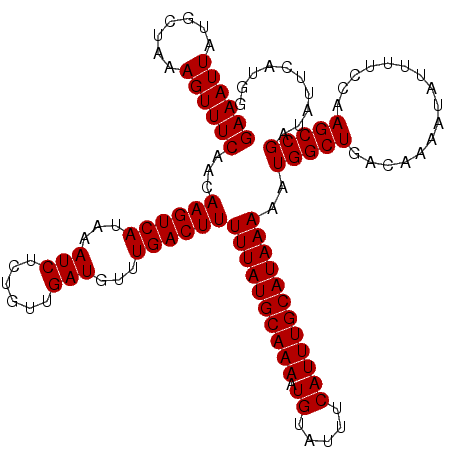
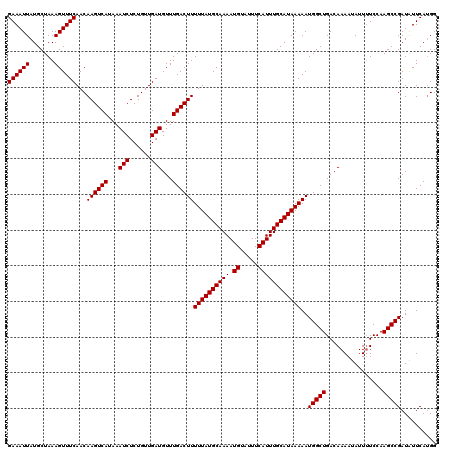
| Location | 1,331,280 – 1,331,400 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.77 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1331280 120 - 20766785 CCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUCAACAGAGAUUUAUGACUUGUUGAAACUUUAGCAUAAUUUC .(((((((.((((((.(.(((....))).).)))).((((((((((((((.....)).))))))))))))..............)).)))))))...((((((....))))))....... ( -26.30) >DroSec_CAF1 49807 120 - 1 CCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUCAACAGAGAUUUAUGACUUGUUGAAACUUUAGCAUAAUUUC .(((((((.((((((.(.(((....))).).)))).((((((((((((((.....)).))))))))))))..............)).)))))))...((((((....))))))....... ( -26.30) >DroEre_CAF1 31463 120 - 1 CCCUGAACAUCGGCUUGGAUAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUCAACAGAGAUUUAUGACUUGUUGAAACUUUAGCAUAAUUUC ...........((((..(((((....))))))))).((((((((((((((.....)).))))))))))))((((.(.(((......))).).)))).((((((....))))))....... ( -26.40) >DroYak_CAF1 16781 120 - 1 CCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUCAACAGAGAUUUAUGACUUGUUGAAACUUUAGCAUAAUUUC .(((((((.((((((.(.(((....))).).)))).((((((((((((((.....)).))))))))))))..............)).)))))))...((((((....))))))....... ( -26.30) >consensus CCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUCAACAGAGAUUUAUGACUUGUUGAAACUUUAGCAUAAUUUC .(((((((.((((((.(.(((....))).).)))).((((((((((((((.....)).))))))))))))..............)).)))))))...((((((....))))))....... (-24.02 = -24.77 + 0.75)

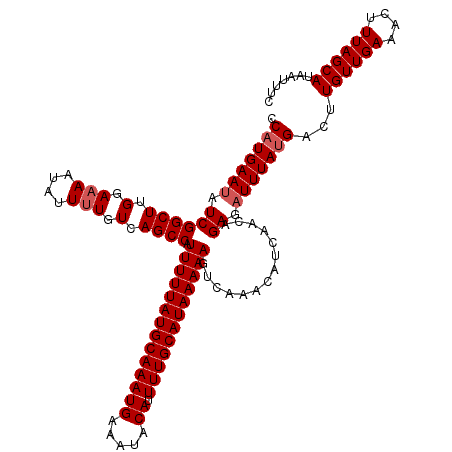
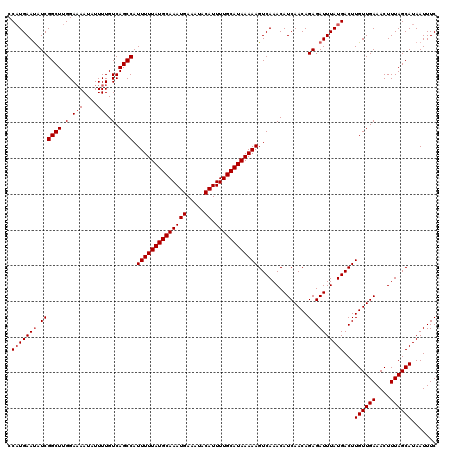
| Location | 1,331,320 – 1,331,428 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.54 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1331320 108 - 20766785 C------CC------CCGCCAGCCGCUGCGCGCCCCACUCCCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUC .------..------.((((((...))).)))...........(((.....((((.(.(((....))).).)))).((((((((((((((.....)).)))))))))))).)))...... ( -22.40) >DroSec_CAF1 49847 114 - 1 C------CCCCGCCCCCGCCAGCCGCUGGGCGCCCCACUCCCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUC .------...(((((..((.....)).)))))...........(((.....((((.(.(((....))).).)))).((((((((((((((.....)).)))))))))))).)))...... ( -27.50) >DroEre_CAF1 31503 120 - 1 CCCACGCCCCCUCCCCCACCAGCCGCCGCACGCCCCACUACCCUGAACAUCGGCUUGGAUAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUC .....................((....))..............(((.....((((..(((((....))))))))).((((((((((((((.....)).)))))))))))).)))...... ( -21.00) >DroYak_CAF1 16821 108 - 1 C------------CCCCACCAUCCGCCGCACGCCCCAUUCCCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUC .------------.....(((...((((.......(((....))).....)))).))).........(((......((((((((((((((.....)).)))))))))))).....))).. ( -18.90) >consensus C______CC____CCCCACCAGCCGCCGCACGCCCCACUCCCAUGAAUAUCGGCUUGGAAAAUAUUUUGUCAGCCAUUUUUAUGCAAAUGAAAUACAUUUUGCAUAAAAAGUCAAACAUC ..................((((((((.....))..((......))......))).))).........(((......((((((((((((((.....)).)))))))))))).....))).. (-17.45 = -17.70 + 0.25)

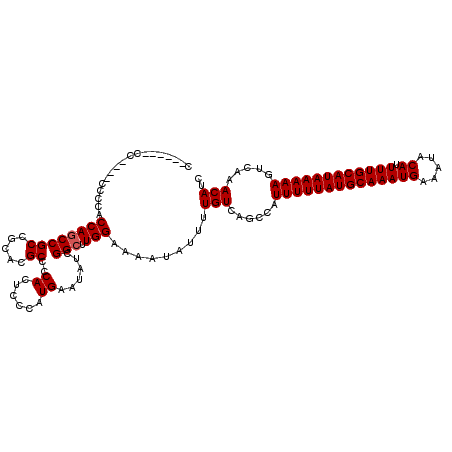
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:05 2006