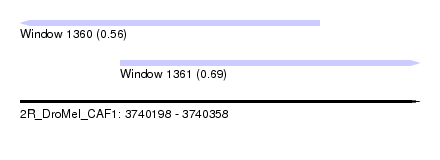
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,740,198 – 3,740,358 |
| Length | 160 |
| Max. P | 0.692010 |

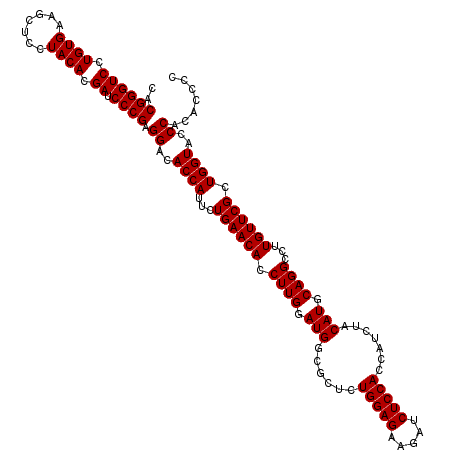
| Location | 3,740,198 – 3,740,318 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -34.65 |
| Energy contribution | -34.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3740198 120 - 20766785 CACGGGUCCUGUGAAGCUCCUACACGAUCCCGAGGACACCAUUCUGAACACCUUGGAUGGCGCUCUGGAGAAGAUCUCCACCAUCUACAUGCAGGCCUUGUUCGCUGGUACCCACACCCC ..((((((.((((.......)))).)).)))).((..((((...((((((((((((((((.....(((((.....)))))))))))).....)))...)))))).))))..))....... ( -36.70) >DroSim_CAF1 2905 120 - 1 CACGGGUCCUGUGAAGCUCCUACACGAUCCCGCGGACACCAUUCUGAACAGCUUGGAUGUCGCUCUGGAGAAGAUCUCCACCAUCUACAUGCAGGCCUUGUUCGCUGGUACCCACACCCC ..((((((.((((.......)))).)).)))).((..((((...(((((((((((.((((.(...(((((.....)))))....).)))).)))))..)))))).))))..))....... ( -38.70) >DroYak_CAF1 2826 120 - 1 CACGGGUCCUGUGAAACUCCUACACGAUCCCGAGGACACCAUUCUGAACACCUUGGAUGGCGCUCUGGAGAAGAUCUCCACUAUCUACAUGCAGGCCUUGUUCGCUGGUACCCACACACC ..((((((.((((.......)))).)).)))).((..((((...((((((.((((.(((......(((((.....))))).......))).))))...)))))).))))..))....... ( -35.22) >consensus CACGGGUCCUGUGAAGCUCCUACACGAUCCCGAGGACACCAUUCUGAACACCUUGGAUGGCGCUCUGGAGAAGAUCUCCACCAUCUACAUGCAGGCCUUGUUCGCUGGUACCCACACCCC ..((((((.((((.......)))).)).)))).((..((((...((((((.((((.(((......(((((.....))))).......))).))))...)))))).))))..))....... (-34.65 = -34.65 + 0.00)

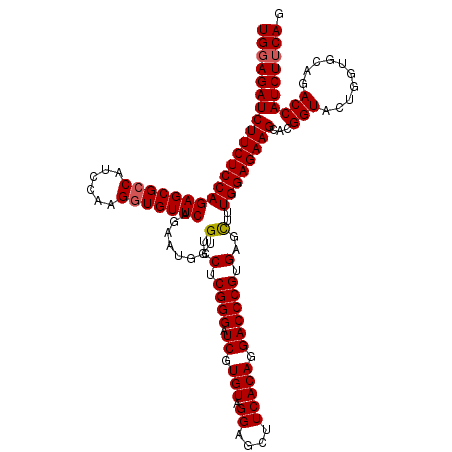
| Location | 3,740,238 – 3,740,358 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -48.27 |
| Consensus MFE | -38.69 |
| Energy contribution | -39.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3740238 120 + 20766785 UGGAGAUCUUCUCCAGAGCGCCAUCCAAGGUGUUCAGAAUGGUGUCCUCGGGAUCGUGUAGGAGCUUCACAGGACCCGUGAGCUUUGGAGAAGCACGGUAAUGGUGCAGACCAUCUUCAG (((((((.((((((((((((((......)))))))........(..(.((((.((.(((.((....))))).)))))).)..)..)))))))((((.......)))).....))))))). ( -44.70) >DroSim_CAF1 2945 120 + 1 UGGAGAUCUUCUCCAGAGCGACAUCCAAGCUGUUCAGAAUGGUGUCCGCGGGAUCGUGUAGGAGCUUCACAGGACCCGUGAGCUUUGGAGAAGCACGGUACUGGCGCAGACCAUCUUCAG (((((((((((((((((((((((.(((..((....))..)))))))((((((.((.(((.((....))))).)))))))).))))))))))))...(((.(((...))))))))))))). ( -54.70) >DroYak_CAF1 2866 120 + 1 UGGAGAUCUUCUCCAGAGCGCCAUCCAAGGUGUUCAGAAUGGUGUCCUCGGGAUCGUGUAGGAGUUUCACAGGACCCGUGAGUUUUGGAGAAGCACGGUACUUGUGCAUACCAUCUGCAG (((((.....)))))(((((((......)))))))(((.((((((.(.(((((((((((.....((((.((((((......)))))))))).))))))).)))).).))))))))).... ( -45.40) >consensus UGGAGAUCUUCUCCAGAGCGCCAUCCAAGGUGUUCAGAAUGGUGUCCUCGGGAUCGUGUAGGAGCUUCACAGGACCCGUGAGCUUUGGAGAAGCACGGUACUGGUGCAGACCAUCUUCAG ((((((((((((((((((((((......)))))))........(..(.((((.((.(((.((....))))).)))))).)..)..))))))))...(((..........)))))))))). (-38.69 = -39.13 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:04 2006