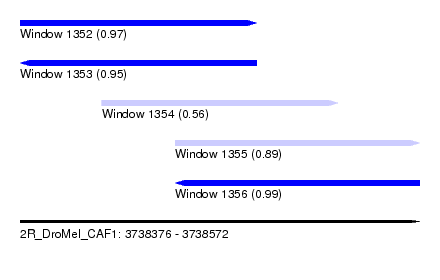
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,738,376 – 3,738,572 |
| Length | 196 |
| Max. P | 0.990590 |

| Location | 3,738,376 – 3,738,492 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3738376 116 + 20766785 UACACUUGUUACGCACAAGAAAUCCAAAUCGGCCCAAGAAACGCAGAUAGCUGGGUCCAUUCAGGACCU----AUUUUUGUUUUGGCUGUGAGAAACGCAGCCGAUUUGAAUUUGCCGAU ....(((((.....)))))........((((((.((((((..((.....))(((((((.....))))))----)))))))..(((((((((.....))))))))).........)))))) ( -39.20) >DroSim_CAF1 1136 119 + 1 UACACUUGUUACCCACAAGAAAUCCAAAUCGGCCCAAGAAACGCAGAUAGCUGGGUCCAUUCAGGACCUACAUAUUUUUGU-UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAU ....(((((.....)))))........((((((.....(((((.(((((..(((((((.....)))))))..))))).)))-))((((((.......))))))...........)))))) ( -38.60) >DroYak_CAF1 1129 115 + 1 UACACUUGUUACCCACAAGAAAUCCAAAUCGGCCCAAGAAACGCAGAUAGCUGGGUCCAUUCAGGACCU----AUUUCGGU-UUGGCUGUGAGAAAUGCAGCCGAUUUGAAUUUGCCGAU ....(((((.....)))))........((((((.........((.....))(((((((.....))))))----).(((((.-((((((((.......)))))))).)))))...)))))) ( -37.50) >consensus UACACUUGUUACCCACAAGAAAUCCAAAUCGGCCCAAGAAACGCAGAUAGCUGGGUCCAUUCAGGACCU____AUUUUUGU_UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAU ....(((((.....)))))........((((((.........(((((.....((((((.....)))))).......))))).((((((((.......)))))))).........)))))) (-34.22 = -34.33 + 0.11)

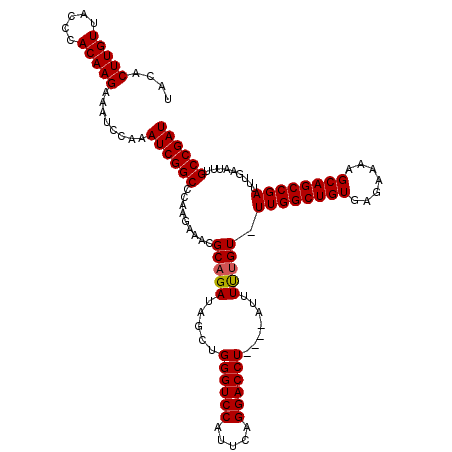
| Location | 3,738,376 – 3,738,492 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -34.73 |
| Energy contribution | -34.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3738376 116 - 20766785 AUCGGCAAAUUCAAAUCGGCUGCGUUUCUCACAGCCAAAACAAAAAU----AGGUCCUGAAUGGACCCAGCUAUCUGCGUUUCUUGGGCCGAUUUGGAUUUCUUGUGCGUAACAAGUGUA ....((((((((((((((((((.(.....).)).(((((((......----.(((((.....)))))..((.....)))))..))))))))))))))))).(((((.....)))))))). ( -36.30) >DroSim_CAF1 1136 119 - 1 AUCGGCAAAUUCAAAUCGGCUGCUUUUCUCACAGCCAA-ACAAAAAUAUGUAGGUCCUGAAUGGACCCAGCUAUCUGCGUUUCUUGGGCCGAUUUGGAUUUCUUGUGGGUAACAAGUGUA ....((((((((((((((((((((........))).((-((...........(((((.....)))))..((.....))))))....)))))))))))))).(((((.....)))))))). ( -36.60) >DroYak_CAF1 1129 115 - 1 AUCGGCAAAUUCAAAUCGGCUGCAUUUCUCACAGCCAA-ACCGAAAU----AGGUCCUGAAUGGACCCAGCUAUCUGCGUUUCUUGGGCCGAUUUGGAUUUCUUGUGGGUAACAAGUGUA ....(((((((((((((((((((..........))...-...(((((----.(((((.....)))))..((.....)))))))...)))))))))))))).(((((.....)))))))). ( -36.70) >consensus AUCGGCAAAUUCAAAUCGGCUGCAUUUCUCACAGCCAA_ACAAAAAU____AGGUCCUGAAUGGACCCAGCUAUCUGCGUUUCUUGGGCCGAUUUGGAUUUCUUGUGGGUAACAAGUGUA ....(((((((((((((((((((..........))(((.((...........(((((.....)))))..((.....))))...))))))))))))))))).(((((.....)))))))). (-34.73 = -34.73 + -0.00)

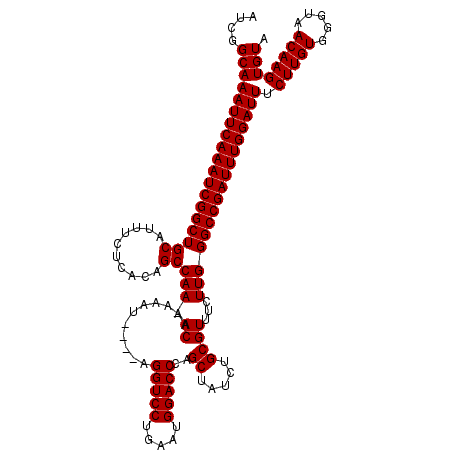
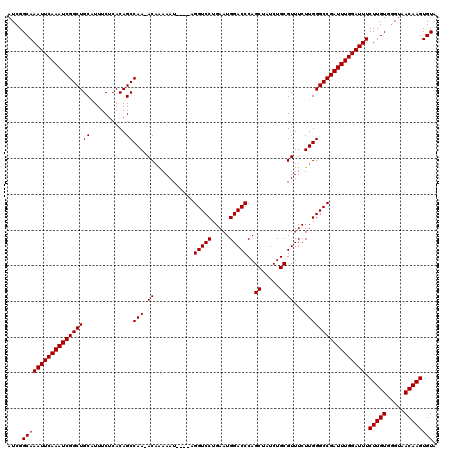
| Location | 3,738,416 – 3,738,532 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -33.70 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3738416 116 + 20766785 ACGCAGAUAGCUGGGUCCAUUCAGGACCU----AUUUUUGUUUUGGCUGUGAGAAACGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGC ..((((((...(((((((.....))))))----)...(((..(((((((((.....)))))))))..)))))))))...((((.(((.....(((((.......)))))..))))))).. ( -37.60) >DroSim_CAF1 1176 119 + 1 ACGCAGAUAGCUGGGUCCAUUCAGGACCUACAUAUUUUUGU-UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGC (((.(((((..(((((((.....)))))))..))))).)))-...((((..(((((((.....((..((.(((..(((.(((....)))..)))))).))..))...)))))))..)))) ( -36.80) >DroYak_CAF1 1169 115 + 1 ACGCAGAUAGCUGGGUCCAUUCAGGACCU----AUUUCGGU-UUGGCUGUGAGAAAUGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGC ..((((((...(((((((.....))))))----)..((((.-((((((((.......)))))))).))))))))))...((((.(((.....(((((.......)))))..))))))).. ( -37.70) >consensus ACGCAGAUAGCUGGGUCCAUUCAGGACCU____AUUUUUGU_UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGC ..((((((....((((((.....)))))).............((((((((.......)))))))).....))))))...((((.(((.....(((((.......)))))..))))))).. (-33.70 = -33.70 + -0.00)

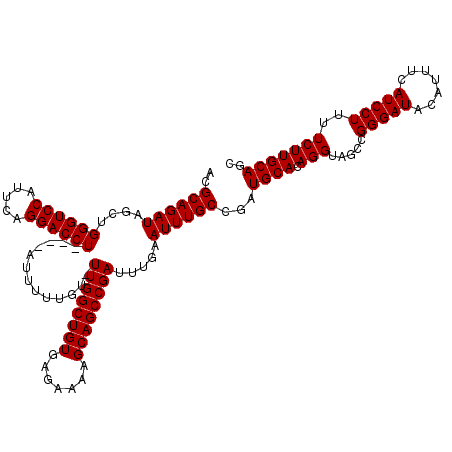
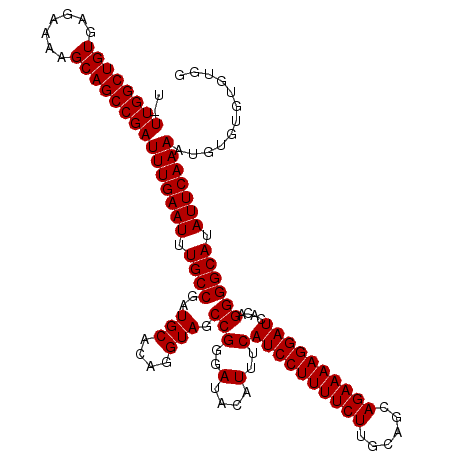
| Location | 3,738,452 – 3,738,572 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -37.47 |
| Energy contribution | -37.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3738452 120 + 20766785 UUUUGGCUGUGAGAAACGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGCAGAAAAGGAUCACAGGGGCAUAUUCAAAAUGUGUGUGUGG ..(((((((((.....)))))))))(((((((.((((..(((....))).(((..(....)..)((((((((((......))))))))))....)))))).)))))))............ ( -39.50) >DroSim_CAF1 1216 119 + 1 U-UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGCAGAAAAGGAUCACAGGGGCAUAUUCAAAAUGUGUGUGUGG .-((((((((.......))))))))(((((((.((((..(((....))).(((..(....)..)((((((((((......))))))))))....)))))).)))))))............ ( -37.50) >DroYak_CAF1 1205 119 + 1 U-UUGGCUGUGAGAAAUGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGCAGAAAAGGAUCACAGGGGCAUAUUCAAAAUGUGUGUGUGG .-((((((((.......))))))))(((((((.((((..(((....))).(((..(....)..)((((((((((......))))))))))....)))))).)))))))............ ( -37.40) >consensus U_UUGGCUGUGAGAAAAGCAGCCGAUUUGAAUUUGCCGAUGCACAGGUAGCCGGGAUACAUUUCAUCCUUUUCUUGCAGCAGAAAAGGAUCACAGGGGCAUAUUCAAAAUGUGUGUGUGG ..((((((((.......))))))))(((((((.((((..(((....))).(((..(....)..)((((((((((......))))))))))....)))))).)))))))............ (-37.47 = -37.47 + -0.00)

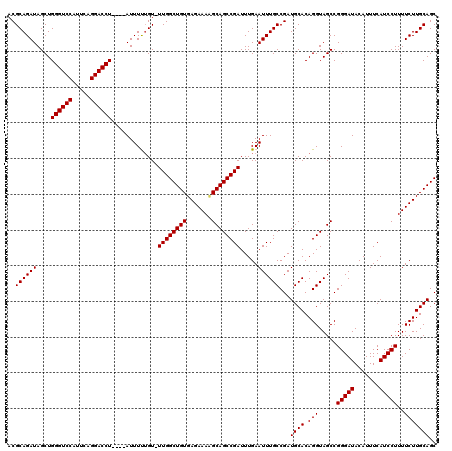
| Location | 3,738,452 – 3,738,572 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -35.00 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3738452 120 - 20766785 CCACACACACAUUUUGAAUAUGCCCCUGUGAUCCUUUUCUGCUGCAAGAAAAGGAUGAAAUGUAUCCCGGCUACCUGUGCAUCGGCAAAUUCAAAUCGGCUGCGUUUCUCACAGCCAAAA ............(((((((.((((....(.((((((((((......)))))))))).).((((((...((...)).)))))).)))).)))))))..(((((.(.....).))))).... ( -36.60) >DroSim_CAF1 1216 119 - 1 CCACACACACAUUUUGAAUAUGCCCCUGUGAUCCUUUUCUGCUGCAAGAAAAGGAUGAAAUGUAUCCCGGCUACCUGUGCAUCGGCAAAUUCAAAUCGGCUGCUUUUCUCACAGCCAA-A ............(((((((.((((....(.((((((((((......)))))))))).).((((((...((...)).)))))).)))).)))))))..(((((.........)))))..-. ( -35.00) >DroYak_CAF1 1205 119 - 1 CCACACACACAUUUUGAAUAUGCCCCUGUGAUCCUUUUCUGCUGCAAGAAAAGGAUGAAAUGUAUCCCGGCUACCUGUGCAUCGGCAAAUUCAAAUCGGCUGCAUUUCUCACAGCCAA-A ............(((((((.((((....(.((((((((((......)))))))))).).((((((...((...)).)))))).)))).)))))))..(((((.........)))))..-. ( -35.00) >consensus CCACACACACAUUUUGAAUAUGCCCCUGUGAUCCUUUUCUGCUGCAAGAAAAGGAUGAAAUGUAUCCCGGCUACCUGUGCAUCGGCAAAUUCAAAUCGGCUGCAUUUCUCACAGCCAA_A ............(((((((.((((....(.((((((((((......)))))))))).).((((((...((...)).)))))).)))).)))))))..(((((.........))))).... (-35.00 = -35.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:59 2006