
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
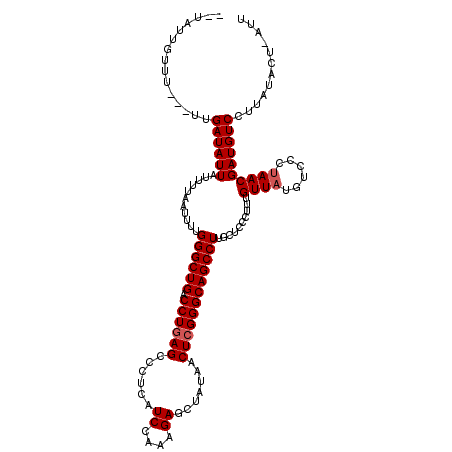
| Location | 3,737,387 – 3,737,501 |
| Length | 114 |
| Max. P | 0.766066 |

| Location | 3,737,387 – 3,737,501 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -21.05 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
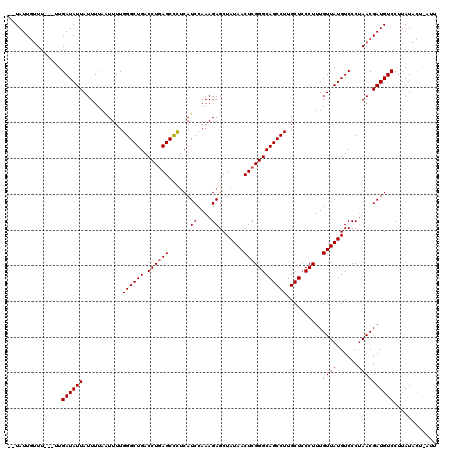
>2R_DroMel_CAF1 3737387 114 + 20766785 --UAUUGUUU---UUGAUAUUAUUUUAGUUUUGGGCUGACCUGAGCCCUCAUCCAAAGAGCUAUAACUCGGGCAGCCUUGCUCCCUUUGUUAUGUCCCUAACGAUGUCCUUAUACU-AUU --(((((((.---..(((((.((...(((...((((((.((((((..(((.......)))......)))))))))))).)))......)).)))))...)))))))..........-... ( -28.50) >DroSim_CAF1 156 114 + 1 --UAUUGUUU---UUGAUAUUAUUUUAAUUUUGGGCUGACCUGAGCCUUCAUCCAAAGAGCUAUAACUCGGGCAGCCUUGCUCCCUUUGUUAUGUCCCUAACGAUGUCCUUAUACU-CUU --(((((((.---..(((((............((((((.((((((......((....)).......)))))))))))).............)))))...)))))))..........-... ( -26.73) >DroYak_CAF1 158 120 + 1 GAUAUUUCUUUAAUUGAUAUUAUUUAAAUUUUGGGCUGACCUCAGCUCCCAUCCAAAGAGCUAUAACUCGGGCAGCCUUGCUACCUUUGUUAUGUCCCCAACGAUGUCCUUAUAUUUUUU ((((((.......((((......))))...(((((..(((...(((((.........)))))(((((..(((.(((...))).)))..))))))))))))).))))))............ ( -24.50) >consensus __UAUUGUUU___UUGAUAUUAUUUUAAUUUUGGGCUGACCUGAGCCCUCAUCCAAAGAGCUAUAACUCGGGCAGCCUUGCUCCCUUUGUUAUGUCCCUAACGAUGUCCUUAUACU_AUU ...............((((((...........((((((.((((((......((....)).......))))))))))))..........((((......))))))))))............ (-21.05 = -21.72 + 0.67)

| Location | 3,737,387 – 3,737,501 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
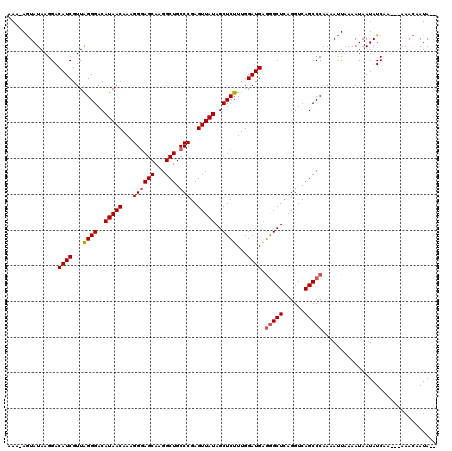
>2R_DroMel_CAF1 3737387 114 - 20766785 AAU-AGUAUAAGGACAUCGUUAGGGACAUAACAAAGGGAGCAAGGCUGCCCGAGUUAUAGCUCUUUGGAUGAGGGCUCAGGUCAGCCCAAAACUAAAAUAAUAUCAA---AAACAAUA-- ..(-(((.......((((...((((..(((((...((((((...))).)))..)))))..))))...)))).(((((......)))))...))))............---........-- ( -30.30) >DroSim_CAF1 156 114 - 1 AAG-AGUAUAAGGACAUCGUUAGGGACAUAACAAAGGGAGCAAGGCUGCCCGAGUUAUAGCUCUUUGGAUGAAGGCUCAGGUCAGCCCAAAAUUAAAAUAAUAUCAA---AAACAAUA-- ..(-(.(((.....((((...((((..(((((...((((((...))).)))..)))))..))))...))))..((((......))))..........)))...))..---........-- ( -24.70) >DroYak_CAF1 158 120 - 1 AAAAAAUAUAAGGACAUCGUUGGGGACAUAACAAAGGUAGCAAGGCUGCCCGAGUUAUAGCUCUUUGGAUGGGAGCUGAGGUCAGCCCAAAAUUUAAAUAAUAUCAAUUAAAGAAAUAUC ..............((((...((((..(((((...((((((...))))))...)))))..))))...))))((.((((....))))))................................ ( -29.10) >consensus AAA_AGUAUAAGGACAUCGUUAGGGACAUAACAAAGGGAGCAAGGCUGCCCGAGUUAUAGCUCUUUGGAUGAGGGCUCAGGUCAGCCCAAAAUUAAAAUAAUAUCAA___AAACAAUA__ ..............((((...((((..(((((...((((((...))).)))..)))))..))))...)))).(((((......)))))................................ (-21.79 = -22.57 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:54 2006