
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
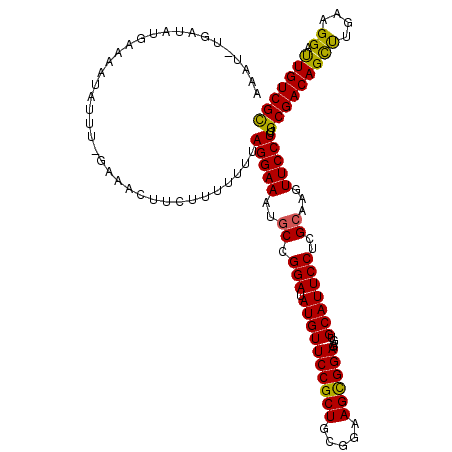
| Location | 3,730,556 – 3,730,671 |
| Length | 115 |
| Max. P | 0.842147 |

| Location | 3,730,556 – 3,730,671 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -28.44 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
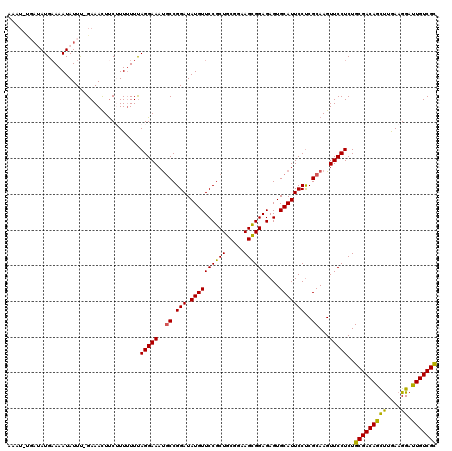
>2R_DroMel_CAF1 3730556 115 + 20766785 G----UGAUAUGUAAAUAUUU-GAAACUUCGUUUUUUAGGAAAUGCCGGAUAUGUUCCGCUGCGGAAGCGGAGAGUGCAUUCCUCGCAAGUUCCUCUGCGACAGCUUGAAGGAUUGUCGU .----.(((((....))))).-...............(((((.(((.(((.((((.((.((((....)))).).).)))))))..)))..)))))..(((((((((....)).))))))) ( -30.60) >DroSec_CAF1 8353 118 + 1 AAAUAUGAUAUGAAAAUAUUU-GAAACUUCUUU-UUUAGGAAAUGCCGGAUAUGUUCCGCUGCGGAAGCGGAGAGUGCAUUCCUCGCAAGUUCCUCUGCGACAGUUUGAAGGAUUGUCGC ......(((((....))))).-...........-...(((((.(((.(((.((((.((.((((....)))).).).)))))))..)))..)))))..(((((((((.....))))))))) ( -35.20) >DroSim_CAF1 8651 115 + 1 GAAU----UAUGAAAAUAUUU-GAAACUUCUUUUUUUAGGAAAUGCCGGAUAUGUUCCGCUGCGGAAGCGGAGAGUGCAUUCCUCGCAAGUUCCUCUGCGACAGUUUGAAGGAUUGUCGC ....----.............-...............(((((.(((.(((.((((.((.((((....)))).).).)))))))..)))..)))))..(((((((((.....))))))))) ( -33.30) >DroEre_CAF1 8844 119 + 1 -AAUUCGAAAUGAAAAUAUUCCGAAAUUCCUUUUUUCAGGAAAUGCCGGAUAUGUUCCGCUGCGGAAGUGGAGAGUGCAUUCCCCGCAAGUUCCUCUGCGACAGCCUAAAGGACUGUCGC -..((((.((((....)))).))))............(((((.(((.((..((((((((((.....))))))....))))..)).)))..)))))..(((((((((....)).))))))) ( -37.50) >DroYak_CAF1 8103 119 + 1 AGAUUUCAAAUGAAAAUAUUC-GAAACUUCGUUUUUUAGGAAACUCCGGAUAUGUUCCGCUGCGGAAGUGGAGAGUGCAUUCCUCGCAAGUUCCUCUGCGACAGCUUAAAGGACUGUCGC (((((((....))))......-(.(((((((......(((((((((((((.....))))(..(....)..).))))...))))))).))))).))))(((((((((....)).))))))) ( -32.40) >consensus AAAU_UGAUAUGAAAAUAUUU_GAAACUUCUUUUUUUAGGAAAUGCCGGAUAUGUUCCGCUGCGGAAGCGGAGAGUGCAUUCCUCGCAAGUUCCUCUGCGACAGCUUGAAGGAUUGUCGC .....................................(((((..((.(((.((((((((((.....))))))....)))))))..))...)))))..(((((((((....)).))))))) (-28.44 = -27.60 + -0.84)

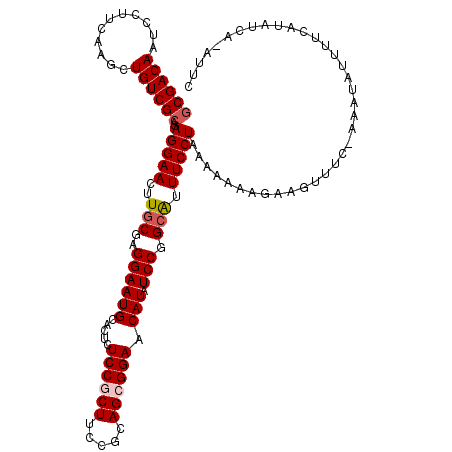
| Location | 3,730,556 – 3,730,671 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -23.52 |
| Energy contribution | -24.16 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.842147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
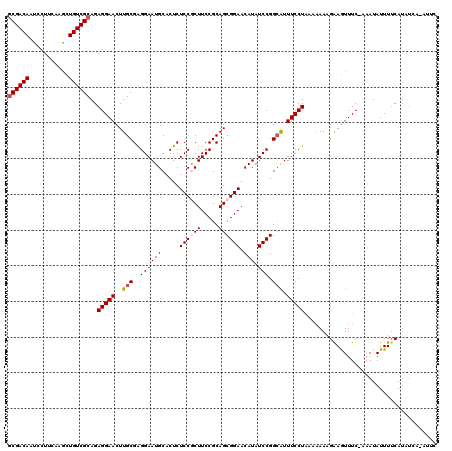
>2R_DroMel_CAF1 3730556 115 - 20766785 ACGACAAUCCUUCAAGCUGUCGCAGAGGAACUUGCGAGGAAUGCACUCUCCGCUUCCGCAGCGGAACAUAUCCGGCAUUUCCUAAAAAACGAAGUUUC-AAAUAUUUACAUAUCA----C .(((((...........)))))....(((((((.((((((((((....((((((.....)))))).(......)))).)))))......)))))))))-................----. ( -25.80) >DroSec_CAF1 8353 118 - 1 GCGACAAUCCUUCAAACUGUCGCAGAGGAACUUGCGAGGAAUGCACUCUCCGCUUCCGCAGCGGAACAUAUCCGGCAUUUCCUAAA-AAAGAAGUUUC-AAAUAUUUUCAUAUCAUAUUU ((((((...........))))))...(((((((.(.((((((((....((((((.....)))))).(......)))).)))))...-...))))))))-..................... ( -29.30) >DroSim_CAF1 8651 115 - 1 GCGACAAUCCUUCAAACUGUCGCAGAGGAACUUGCGAGGAAUGCACUCUCCGCUUCCGCAGCGGAACAUAUCCGGCAUUUCCUAAAAAAAGAAGUUUC-AAAUAUUUUCAUA----AUUC ((((((...........))))))..(((((..(((..((((((.....((((((.....)))))).))).))).))).))))).......((((....-......))))...----.... ( -29.10) >DroEre_CAF1 8844 119 - 1 GCGACAGUCCUUUAGGCUGUCGCAGAGGAACUUGCGGGGAAUGCACUCUCCACUUCCGCAGCGGAACAUAUCCGGCAUUUCCUGAAAAAAGGAAUUUCGGAAUAUUUUCAUUUCGAAUU- (((((((((.....))))))))).......(((.((((((((((.........(((((...)))))........))))))))))....)))....((((((((......))))))))..- ( -41.63) >DroYak_CAF1 8103 119 - 1 GCGACAGUCCUUUAAGCUGUCGCAGAGGAACUUGCGAGGAAUGCACUCUCCACUUCCGCAGCGGAACAUAUCCGGAGUUUCCUAAAAAACGAAGUUUC-GAAUAUUUUCAUUUGAAAUCU ((((((((.......))))))))((.((((((((((.(((.((.......)).))))))..((((.....)))))))))))))..........(((((-((((......))))))))).. ( -34.90) >consensus GCGACAAUCCUUCAAGCUGUCGCAGAGGAACUUGCGAGGAAUGCACUCUCCGCUUCCGCAGCGGAACAUAUCCGGCAUUUCCUAAAAAAAGAAGUUUC_AAAUAUUUUCAUAUCA_AUUC ((((((...........))))))..(((((..(((..((((((.....((((((.....)))))).))).))).))).)))))..................................... (-23.52 = -24.16 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:51 2006