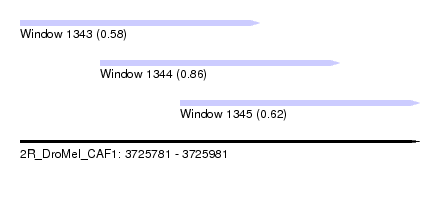
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,725,781 – 3,725,981 |
| Length | 200 |
| Max. P | 0.858746 |

| Location | 3,725,781 – 3,725,901 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -45.32 |
| Consensus MFE | -26.50 |
| Energy contribution | -25.84 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3725781 120 + 20766785 AUAACACUCCGGAGGGAUAUAUCUGCUCCUGUCCAGACCAUCUAUACCUGCAGCCAAACGGAAAGCGCUGCAGCAUGCAGCACGCCUGCGAUCACUGGGAUACGUGUUCACAGGUGUGCG .......((.(((((((.........)))).))).))..........((((((((.........).)))))))......(((((((((.((.(((........))).)).))))))))). ( -39.70) >DroVir_CAF1 3646 120 + 1 ACAAUACGCCCGAGGGUUACAUCUGUUCAUGCCCGCCACAUUUGUAUUUGCAGCCGAAUGGCAAACGCUGCAGCAUGCAGCACGCUUGUGAGCAUUGGGACACCUGCUCACAGGUGUGCG .............((((.((....))....))))........((((((((((((............))))))).)))))(((((((((((((((..((....))))))))))))))))). ( -48.10) >DroGri_CAF1 3974 120 + 1 ACAACACGCCCGAGGGUUACAUCUGCUCGUGUCCCCCACAUUUGUAUUUGCAGCCAAAUGGCAAGCGUUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACGUGUGCCCAGGUCUGUG ...(((.(((...((((.......((.((((((((.......((((..(((.(((....)))..))).))))((.(((((.....))))).))...)))))))).)))))).))).))). ( -43.61) >DroWil_CAF1 3259 120 + 1 AUAAUACCCCAGAGGGUUAUAUAUGCUCGUGCCCGGCUCACUUGUUCCUGCAACCGAAUGGCAAGCGUUGUAGUAUGCAACAUGCCUGUGAUAAUUGGGAUACAUGCUCACAGGUGUGUG .....((((....))))((((.(((((..(((((((.....((((....)))))))...))))))))))))).......((((((((((((....((.....))...)))))))))))). ( -35.40) >DroMoj_CAF1 4455 120 + 1 ACAAUACGCCCGAGGGCUACAUCUGCUCCUGCCCGCCCCACCUGUAUCUGCAGCCCAAUGGCAAGCGCUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACCUGCUCGCAGGUGUGCG .............((((.............))))........((((((((((((.(........).))))))).)))))(((((((((((((((..((....))))))))))))))))). ( -57.02) >DroAna_CAF1 3249 120 + 1 ACAAUACACCGGAGGGUUACAUCUGCUCCUGCCCGGAUCACUUGUACCUGCAGCCGAACGGGAAGCGGUGCAGCAUGCAGCAUGCCUGCGAUCACUGGGACAGCUGUUCCCAGGUGUGUG ...(((((((...(((..(((.(((.(((((....((((...((((((.((..((.....))..))))))))(((.((.....)).)))))))..)))))))).))).))).))))))). ( -48.10) >consensus ACAAUACGCCCGAGGGUUACAUCUGCUCCUGCCCGCCACACUUGUACCUGCAGCCGAAUGGCAAGCGCUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACCUGCUCACAGGUGUGCG .............((((.............))))........(((..(((((((............)))))))...)))(((((((((.((.((..........)).)).))))))))). (-26.50 = -25.84 + -0.66)

| Location | 3,725,821 – 3,725,941 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -48.68 |
| Consensus MFE | -29.64 |
| Energy contribution | -28.23 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3725821 120 + 20766785 UCUAUACCUGCAGCCAAACGGAAAGCGCUGCAGCAUGCAGCACGCCUGCGAUCACUGGGAUACGUGUUCACAGGUGUGCGAAAGUAGCGGUAAGGGUUAUGACUGCCGUUGCUUGGAUGG .......((((((((.........).)))))))(((.(.(((((((((.((.(((........))).)).)))))))))..(((((((((((.(........))))))))))))).))). ( -50.60) >DroVir_CAF1 3686 120 + 1 UUUGUAUUUGCAGCCGAAUGGCAAACGCUGCAGCAUGCAGCACGCUUGUGAGCAUUGGGACACCUGCUCACAGGUGUGCGAAAACAACGGCAAAGGCUAUGCCUGUCAGUGCCUCGAAGG .((((.(((((.(((....)))....(((((.....)))))(((((((((((((..((....)))))))))))))))))))).)))).((((.((((...)))).....))))....... ( -53.40) >DroGri_CAF1 4014 120 + 1 UUUGUAUUUGCAGCCAAAUGGCAAGCGUUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACGUGUGCCCAGGUCUGUGAGAACAAUGGCAAGAGCUACGCCUGCCAGUGCCUCGAGGG .........((.(((....)))..))(((((.....)))))...(((.((((((((((.(..((((.((.(..((((((....)))..)))..).))))))..).)))))).))))))). ( -43.10) >DroWil_CAF1 3299 120 + 1 CUUGUUCCUGCAACCGAAUGGCAAGCGUUGUAGUAUGCAACAUGCCUGUGAUAAUUGGGAUACAUGCUCACAGGUGUGUGAAACAAAGGGCAAAGGUUAUCAGUGUCGUUGCCUGGAGGG ....((((.(((((.((.((..((.(.((((....((..((((((((((((....((.....))...))))))))))))....))....)))).).))..))...)))))))..)))).. ( -36.70) >DroMoj_CAF1 4495 120 + 1 CCUGUAUCUGCAGCCCAAUGGCAAGCGCUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACCUGCUCGCAGGUGUGCGAGAGCAGCGGCAAGGGCUAUGCCUGCCAGUGCCUGGAGGG ((((((((((((((.(........).))))))).)))).(((((((((((((((..((....))))))))))))))))).........((((..(((.......)))..))))...))). ( -64.70) >DroAna_CAF1 3289 120 + 1 CUUGUACCUGCAGCCGAACGGGAAGCGGUGCAGCAUGCAGCAUGCCUGCGAUCACUGGGACAGCUGUUCCCAGGUGUGUGAGACCAGCGGCAAGGGGUACGAGUGUCGCUGCUUGGAGGG (((((((((...((((...((((.((((.(((((.....)).)))))))..)).(((((((....).))))))..........))..))))...)))))))))................. ( -43.60) >consensus CUUGUACCUGCAGCCGAAUGGCAAGCGCUGCAGCAUGCAGCACGCCUGCGAGCACUGGGACACCUGCUCACAGGUGUGCGAAAACAACGGCAAGGGCUAUGACUGCCAGUGCCUGGAGGG .((((..(((((((............)))))))......(((((((((.((.((..........)).)).)))))))))....)))).((((..(((.......)))..))))....... (-29.64 = -28.23 + -1.41)

| Location | 3,725,861 – 3,725,981 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -45.29 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3725861 120 + 20766785 CACGCCUGCGAUCACUGGGAUACGUGUUCACAGGUGUGCGAAAGUAGCGGUAAGGGUUAUGACUGCCGUUGCUUGGAUGGAUUCGACUUGGGCUUCGAUCGGUUCACAUGCAAAAGCACC ((((((((.((.(((........))).)).))))))))...(((((((((((.(........))))))))))))((((.(((.(((........)))))).))))...(((....))).. ( -42.50) >DroVir_CAF1 3726 120 + 1 CACGCUUGUGAGCAUUGGGACACCUGCUCACAGGUGUGCGAAAACAACGGCAAAGGCUAUGCCUGUCAGUGCCUCGAAGGCUACGAUCUAGCCUACGACAAAUUUACGUGCAAGAGCACA ((((((((((((((..((....))))))))))))))))..........((((.((((...)))).....))))(((.((((((.....)))))).))).........((((....)))). ( -50.90) >DroWil_CAF1 3339 120 + 1 CAUGCCUGUGAUAAUUGGGAUACAUGCUCACAGGUGUGUGAAACAAAGGGCAAAGGUUAUCAGUGUCGUUGCCUGGAGGGUUAUGACUUGGCCUAUGAUAAAUUUACAUGUAAAAGUACC ...((((((((....((.....))...))))))))(((((((........((.((((((.....(((((.(((.....))).))))).)))))).)).....)))))))........... ( -32.22) >DroMoj_CAF1 4535 120 + 1 CACGCCUGCGAGCACUGGGACACCUGCUCGCAGGUGUGCGAGAGCAGCGGCAAGGGCUAUGCCUGCCAGUGCCUGGAGGGCUAUGACCUGGCCUACGACAAGUUCACGUGCAAGAGCACC ((((((((((((((..((....))))))))))))))))...((((...((((..(((.......)))..))))..(.((((((.....)))))).).....))))..((((....)))). ( -58.60) >DroAna_CAF1 3329 120 + 1 CAUGCCUGCGAUCACUGGGACAGCUGUUCCCAGGUGUGUGAGACCAGCGGCAAGGGGUACGAGUGUCGCUGCUUGGAGGGCUUCGACUUGGCCUACGACAAGUUCACCUGCAAGAGCACU ...(((((.((.(((((...))).)).)).)))))((((.....((((((((.(.....)...))))))))((((.(((.....((((((........))))))..))).)))).)))). ( -41.50) >DroPer_CAF1 2960 120 + 1 CAUGCCUGCGACCACUGGGACACCUGCUCGCAGGUGUGCCAGACCAGCGGCAAGGGCUACGACUGUCGCUGUUUGGAGGGAUUCGACCUCGCCUACGACAAAUUCACGUGCAAGAGCACC .......(((((((((((.(((((((....))))))).))))..((((((((.(........)))))))))..))).((.......))))))...............((((....)))). ( -46.00) >consensus CACGCCUGCGAUCACUGGGACACCUGCUCACAGGUGUGCGAAACCAGCGGCAAGGGCUAUGACUGUCGCUGCCUGGAGGGCUACGACCUGGCCUACGACAAAUUCACGUGCAAGAGCACC (((((((((((.((..((....)))).)))))))))))..........((((..(((.......)))..))))..(.(((((.......))))).)............(((....))).. (-24.73 = -24.48 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:48 2006