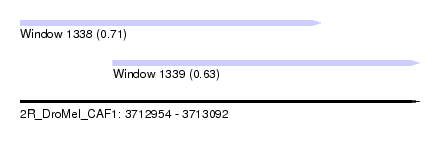
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,712,954 – 3,713,092 |
| Length | 138 |
| Max. P | 0.705614 |

| Location | 3,712,954 – 3,713,058 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.49 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
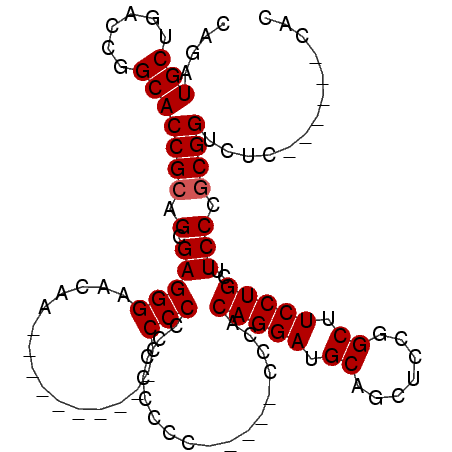
>2R_DroMel_CAF1 3712954 104 + 20766785 CAGAUGCUGACCGGCACCGCAGCGAGGGAACAACACUCAACACCCCCCCCCCCCCCAUUCCCCACAGGAUGCACCUCCGGCUUCCUGCUUCCCGCGGUCUC------CAC .....(((....)))(((((...(.(((((...........................)))))).(((((.((.......)).)))))......)))))...------... ( -23.93) >DroSec_CAF1 36589 89 + 1 CAGAUGCUGACCUGCACCGCAGCGAGGGAACAC----------CCCCUCACCCCC-----CACACAGGAUGCAGCUCCGGCUUCCUGCUUCCCGCGGACUC------CAC ....(((......)))((((...(((((.....----------.)))))......-----....(((((.((.......)).)))))......))))....------... ( -28.30) >DroSim_CAF1 13894 95 + 1 CAGAUGCUGACCGGCACCGCAGCGAGGGAACAA----------CCCCCUCUUUCU-----CCCACAGGAUGCAGCUCCGGCUUCCUGCUUCCCUCGGUCUCCACCUCCAC .((.((..(((((((......))(((((.....----------..))))).....-----....(((((.((.......)).))))).......)))))..)).)).... ( -27.40) >consensus CAGAUGCUGACCGGCACCGCAGCGAGGGAACAA__________CCCCCCCCCCCC_____CCCACAGGAUGCAGCUCCGGCUUCCUGCUUCCCGCGGUCUC______CAC ....(((......)))((((.(.(((((................))).................(((((.((.......)).)))))..))).))))............. (-19.16 = -19.49 + 0.33)

| Location | 3,712,986 – 3,713,092 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
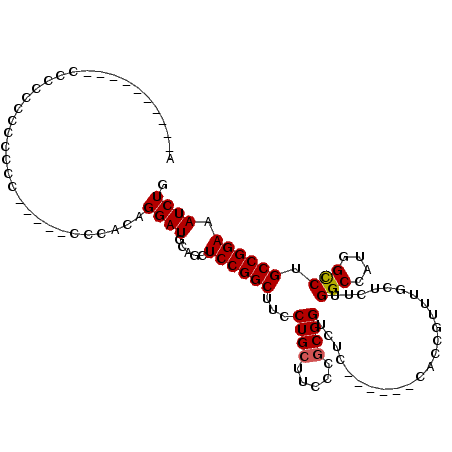
>2R_DroMel_CAF1 3712986 106 + 20766785 ACACUCAACACCCCCCCCCCCCCCAUUCCCCACAGGAUGCACCUCCGGCUUCCUGCUUCCCGCGGUCUC------CACCGUUUGCUUUUGGCCAUGGUCUGCCGGAAAUCUG .......................(((((......)))))....((((((..((.(((....(((((...------.)))))........)))...))...))))))...... ( -22.90) >DroSec_CAF1 36621 91 + 1 C----------CCCCUCACCCCC-----CACACAGGAUGCAGCUCCGGCUUCCUGCUUCCCGCGGACUC------CACAGUUUGCUCUUGGCCAUGGCCUGCCGGAAAUCUG .----------............-----......((((.....((((((............(((((((.------...)))))))....(((....))).)))))).)))). ( -25.30) >DroSim_CAF1 13926 97 + 1 A----------CCCCCUCUUUCU-----CCCACAGGAUGCAGCUCCGGCUUCCUGCUUCCCUCGGUCUCCACCUCCACCGUUUGCUCUUGGCCAUGGCCUGCCGGAAAUCUG .----------...........(-----((....)))..(((.((((((.....((......((((..........))))...))....(((....))).))))))...))) ( -22.50) >consensus A__________CCCCCCCCCCCC_____CCCACAGGAUGCAGCUCCGGCUUCCUGCUUCCCGCGGUCUC______CACCGUUUGCUCUUGGCCAUGGCCUGCCGGAAAUCUG ..................................((((.....((((((...((((.....))))........................(((....))).)))))).)))). (-19.55 = -19.67 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:43 2006