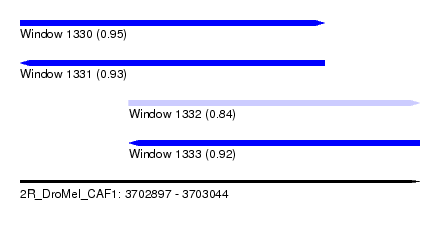
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,702,897 – 3,703,044 |
| Length | 147 |
| Max. P | 0.949405 |

| Location | 3,702,897 – 3,703,009 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.47 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -29.83 |
| Energy contribution | -30.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3702897 112 + 20766785 UCAAAGUGAUUGAUGGUCGAAUGCAUAAAUUAUUGAUGGCCAGCAGACAAUGCUUCAAGUCUGCUGAAGUUGGCCAAAGGCAAGAGCGUUAGUGGGUGAGAGCAUAAAAAUU .......((((.(((.((..((.(((..........((((((((((((..........))))))......))))))((.((....)).)).))).))..)).)))...)))) ( -28.30) >DroSec_CAF1 26385 112 + 1 UCAAAGUGAUUGAUGGUCGAAUGCAUAAAUUAUUGAUGGCCAGCAGACAAUGCUUCAAGUCUGCUGAAGUUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGAGCAUAAAAAUU ......((.(((.((((((....((........)).)))))).))).))((((((((..((..((((((((.(((...)))...))))))))..))))).)))))....... ( -30.70) >DroEre_CAF1 22930 112 + 1 UCAAAGUGAUUGAUGGUCGAAUGCAUAAAUUAUUGAUGGCCAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGACCAUAAAAACU ....(((.....((((((..((.(((..........((((((((((((..........))))))......))))))(((((....))))).))).))..))))))....))) ( -35.40) >DroYak_CAF1 23956 112 + 1 UCAAAGUGAUUGAUGGUCGAAUGCAUAAAUUAUUGAUGGCCAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAAAGCUUUAGUGGGUGAGACCAUAAAAAUU .......((((.((((((..((.(((..........((((((((((((..........))))))......))))))(((((....))))).))).))..))))))...)))) ( -34.50) >consensus UCAAAGUGAUUGAUGGUCGAAUGCAUAAAUUAUUGAUGGCCAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGACCAUAAAAAUU ............((((((..((.(((..........((((((((((((..........))))))......))))))(((((....))))).))).))..))))))....... (-29.83 = -30.57 + 0.75)

| Location | 3,702,897 – 3,703,009 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.47 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3702897 112 - 20766785 AAUUUUUAUGCUCUCACCCACUAACGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUGGCCAUCAAUAAUUUAUGCAUUCGACCAUCAAUCACUUUGA .......((((..............((....))...((((((......((((((..........))))))))))))............)))).......((((.....)))) ( -21.90) >DroSec_CAF1 26385 112 - 1 AAUUUUUAUGCUCUCACCCACUAAAGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUGGCCAUCAAUAAUUUAUGCAUUCGACCAUCAAUCACUUUGA .......((((..............((....))...((((((......((((((..........))))))))))))............)))).......((((.....)))) ( -21.80) >DroEre_CAF1 22930 112 - 1 AGUUUUUAUGGUCUCACCCACUAAAGCUCUUGCCUUUGGCCAUCUUCAGCAGACUUGAAGCAUUGUCUGCUGGCCAUCAAUAAUUUAUGCAUUCGACCAUCAAUCACUUUGA (((..((((((((............((....))...((((((......((((((..........))))))))))))..................)))))).))..))).... ( -26.70) >DroYak_CAF1 23956 112 - 1 AAUUUUUAUGGUCUCACCCACUAAAGCUUUUGCCUUUGGCCAUCUUCAGCAGACUUGAAGCAUUGUCUGCUGGCCAUCAAUAAUUUAUGCAUUCGACCAUCAAUCACUUUGA ........(((((............((....))...((((((......((((((..........))))))))))))..................)))))((((.....)))) ( -26.10) >consensus AAUUUUUAUGCUCUCACCCACUAAAGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUGGCCAUCAAUAAUUUAUGCAUUCGACCAUCAAUCACUUUGA ........(((((............((....))...((((((......((((((..........))))))))))))..................)))))((((.....)))) (-22.88 = -23.37 + 0.50)

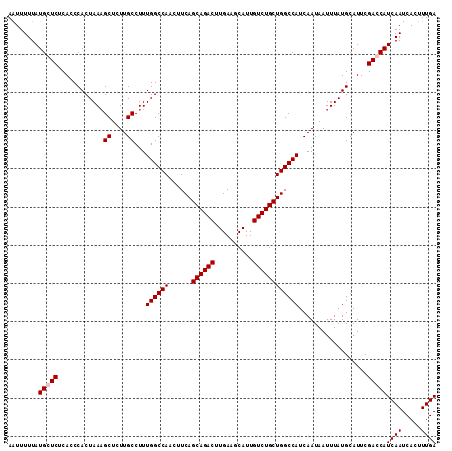
| Location | 3,702,937 – 3,703,044 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.48 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -22.88 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
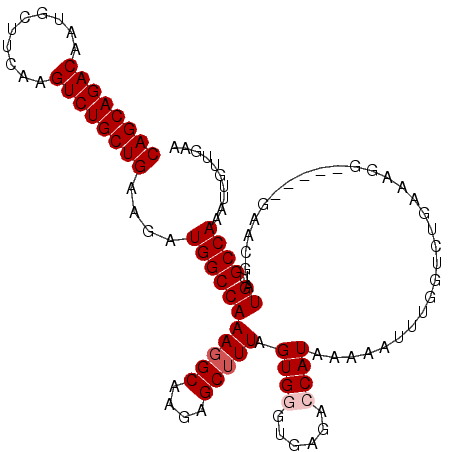
>2R_DroMel_CAF1 3702937 107 + 20766785 CAGCAGACAAUGCUUCAAGUCUGCUGAAGUUGGCCAAAGGCAAGAGCGUUAGUGGGUGAGAGCAUAAAAAUUUGGUCUGAAAGG-----GAACGAUGUGCCAAAUUGUUGAA ((((((((..........))))))))...(..((....((((....((((......(.(((.((........)).))).)....-----.))))...)))).....))..). ( -29.80) >DroSec_CAF1 26425 112 + 1 CAGCAGACAAUGCUUCAAGUCUGCUGAAGUUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGAGCAUAAAAAUUUUGUCUAAAAAGGGAACGAACGAUGUGCCAAAUUGUUGAA ((((((((..........))))))))...(((((((.((((((((..((((....((....)).))))..)))))))).....(....)......)).)))))......... ( -32.10) >DroEre_CAF1 22970 107 + 1 CAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGACCAUAAAAACUUGGUCUGAAUGG-----GAACGAUGUGCCAAAUUGUUGAA ((((((((..........))))))))....(((((((((((....))))).((...(.((((((........)))))).)....-----..))..)).)))).......... ( -31.90) >DroYak_CAF1 23996 105 + 1 CAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAAAGCUUUAGUGGGUGAGACCAUAAAAAUUAGAUCUGAAAGG-----GAA--AUGUGCCAAAUUGUUGAA ((((((((..........))))))))....(((((((((((....))))).((((......))))..........(((....))-----)..--.)).)))).......... ( -28.20) >consensus CAGCAGACAAUGCUUCAAGUCUGCUGAAGAUGGCCAAAGGCAAGAGCUUUAGUGGGUGAGACCAUAAAAAUUUGGUCUGAAAGG_____GAACGAUGUGCCAAAUUGUUGAA ((((((((..........))))))))....(((((((((((....))))).((((......))))..............................)).)))).......... (-22.88 = -23.62 + 0.75)

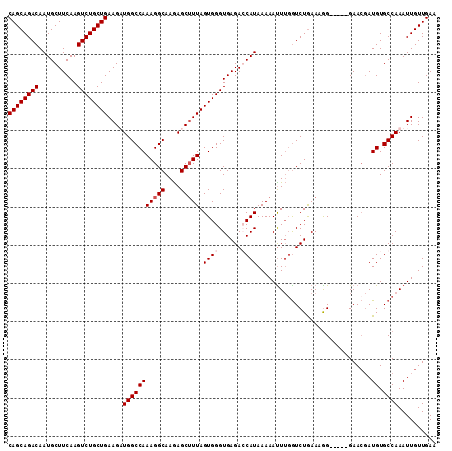
| Location | 3,702,937 – 3,703,044 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.48 |
| Mean single sequence MFE | -22.29 |
| Consensus MFE | -20.54 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
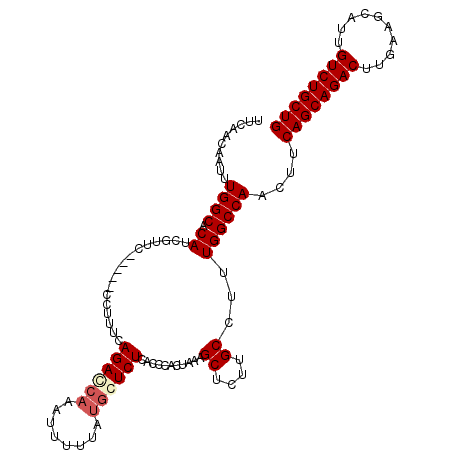
>2R_DroMel_CAF1 3702937 107 - 20766785 UUCAACAAUUUGGCACAUCGUUC-----CCUUUCAGACCAAAUUUUUAUGCUCUCACCCACUAACGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUG .....(((...((((...((((.-----......(((.((........)).)))........))))....)))).)))........((((((((..........)))))))) ( -21.06) >DroSec_CAF1 26425 112 - 1 UUCAACAAUUUGGCACAUCGUUCGUUCCCUUUUUAGACAAAAUUUUUAUGCUCUCACCCACUAAAGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUG .........(((((.........(((.........))).......................((((((....).))))))))))...((((((((..........)))))))) ( -19.60) >DroEre_CAF1 22970 107 - 1 UUCAACAAUUUGGCACAUCGUUC-----CCAUUCAGACCAAGUUUUUAUGGUCUCACCCACUAAAGCUCUUGCCUUUGGCCAUCUUCAGCAGACUUGAAGCAUUGUCUGCUG ..........((((.........-----......((((((........)))))).......((((((....).)))))))))....((((((((..........)))))))) ( -26.20) >DroYak_CAF1 23996 105 - 1 UUCAACAAUUUGGCACAU--UUC-----CCUUUCAGAUCUAAUUUUUAUGGUCUCACCCACUAAAGCUUUUGCCUUUGGCCAUCUUCAGCAGACUUGAAGCAUUGUCUGCUG ..........((((....--...-----......(((((..........))))).......((((((....).)))))))))....((((((((..........)))))))) ( -22.30) >consensus UUCAACAAUUUGGCACAUCGUUC_____CCUUUCAGACCAAAUUUUUAUGCUCUCACCCACUAAAGCUCUUGCCUUUGGCCAACUUCAGCAGACUUGAAGCAUUGUCUGCUG ..........((((.((.................((((((........))))))...........((....))...))))))....((((((((..........)))))))) (-20.54 = -21.48 + 0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:38 2006