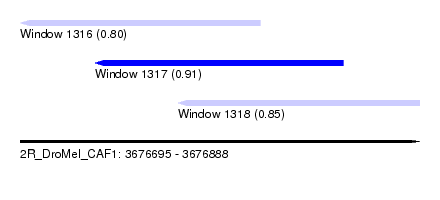
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,676,695 – 3,676,888 |
| Length | 193 |
| Max. P | 0.914080 |

| Location | 3,676,695 – 3,676,811 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3676695 116 - 20766785 CUUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGAAGCAAAGAACUAUUCAAAACUCCACCAGCGCGUGUUUACAAUUUAAAACACACGUGUGGCGU----GUUAG ((((((.(((((..((((((........)))))).))))).....))))))................((.((((..(((.((((((........)))))).))))))).))----..... ( -32.80) >DroSim_CAF1 8955 116 - 1 CCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUAAAAACUACACCAGCGCGUGUUUACAAUUUAAAACACACGUGUGGCUU----GUUAG ((.....))...(((.((((.((((((((((.((((((........)))))).)))))))................(((.((((((........)))))).))).))))))----).))) ( -33.30) >DroEre_CAF1 7914 104 - 1 CCUCAAUGGUGCCUAUCAAGUCUAGGUUCUUGGUUGCGCCCAUUUUUGCAGUAAAGAAC------------CACCAGCGCGUGUUUACAAUUUAAAAUACACGUGUGGAGU----GUUAG .(((..((((..((....))....(((((((..(((((........)))))..))))))------------)))))((((((((..............)))))))).))).----..... ( -28.94) >DroYak_CAF1 5685 116 - 1 CCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUUAAAACUCCACCAGCGCGUGUUUACAAUUUAAAACACACGUGUGGCGU----GUUAG ......((((....))))......(((((((.((((((........)))))).)))))))...(((.((.((((..(((.((((((........)))))).))))))).))----.))). ( -32.70) >DroAna_CAF1 2608 116 - 1 CCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUAUCGUUAAAAAUCAAUGAUCAAAU---GCAGCGCGUGUUUACAAAUCA-AACGCACGUGUGCAGCAGCGGUUAG ....((((((((.(((((((........))))))))))))..(((((....))))))))..(((((...(---((.((((((((..........-...))))))))...)))..))))). ( -32.42) >consensus CCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUAAAAACU_CACCAGCGCGUGUUUACAAUUUAAAACACACGUGUGGCGU____GUUAG .......(((((..((((((........)))))).))))).................................(((.(((((((..............))))))))))............ (-24.34 = -23.70 + -0.64)

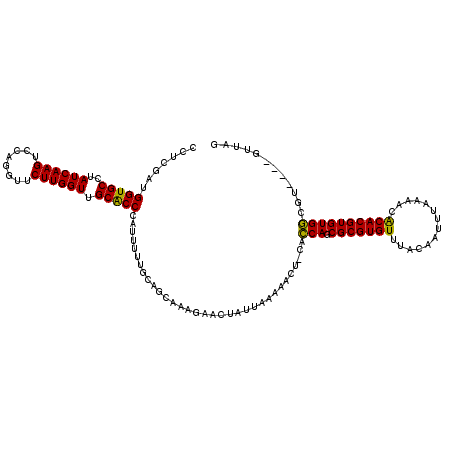
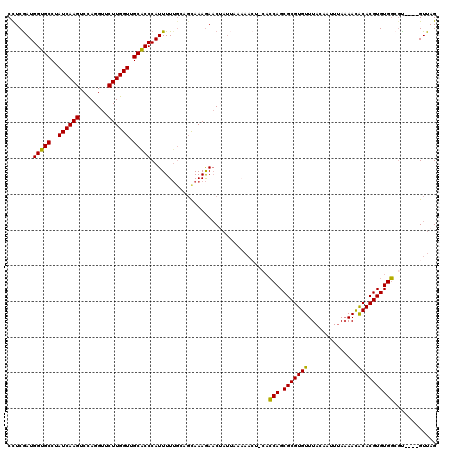
| Location | 3,676,731 – 3,676,851 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.99 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.36 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3676731 120 - 20766785 GCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCUUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGAAGCAAAGAACUAUUCAAAACUCCACCAGCGC ((.(((((....)))))..((((((((.....))....(.((((((.(((((..((((((........)))))).))))).....)))))))..))))))................)).. ( -32.50) >DroSim_CAF1 8991 120 - 1 GCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUAAAAACUACACCAGCGC ((.(((((....)))))..((((.........(((((((((...)))))))))...........(((((((.((((((........)))))).)))))))......))))......)).. ( -37.80) >DroEre_CAF1 7950 108 - 1 GCAUCGGAGUCCUCCGACAAGUUCUCGACCUCGGCAUCGUCCUCAAUGGUGCCUAUCAAGUCUAGGUUCUUGGUUGCGCCCAUUUUUGCAGUAAAGAAC------------CACCAGCGC ((.(((((....))))).........(((...((((((((.....))))))))......)))..(((((((..(((((........)))))..))))))------------)....)).. ( -34.40) >DroYak_CAF1 5721 120 - 1 GCAUCGGAGUCCUCCGACAAGUUCUCGACCUCGGCAUCGUCCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUUAAAACUCCACCAGCGC ((...(((((.....(((........(....)(((((((((...)))))))))......)))..(((((((.((((((........)))))).))))))).......)))))....)).. ( -37.70) >DroAna_CAF1 2647 117 - 1 GCAUCGGAGUCCUCUGAAAGGUUCUCUACCUCUGCAUCAUCCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUAUCGUUAAAAAUCAAUGAUCAAAU---GCAGCGC ((.(((((....))))).((((.....))))((((............(((((.(((((((........)))))))))))).((((.((((((.......)))))).))))---)))).)) ( -31.00) >consensus GCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACCCAUUUUUGCAGCAAAGAACUAUUAAAAACU_CACCAGCGC ((.(((((....)))))...............(((((((((...)))))))))............((((((.((((((........)))))).)))))).................)).. (-26.60 = -27.36 + 0.76)

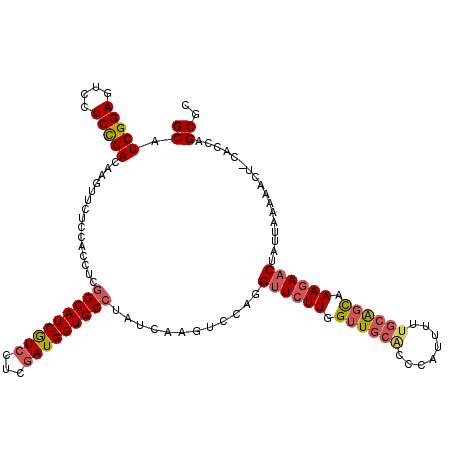
| Location | 3,676,771 – 3,676,888 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.21 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.66 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3676771 117 - 20766785 UCGUUUUUAAAAGUGUGAG-CAACCUACCUCGACCUCCGCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCUUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACC ..................(-(((((.((((.(((.........((((..((......))..)))).....(((((((((...)))))))))......))).))))....))))))... ( -38.80) >DroSim_CAF1 9031 117 - 1 UCGUUUUUAAAAGUGCGAG-CUACCUACCUCGACCUCCGCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACC ............((((.((-(((...((((.(((.........((((..((......))..)))).....(((((((((...)))))))))......))).))))...))))))))). ( -38.10) >DroEre_CAF1 7978 116 - 1 UCGGU-GUAAAAGUGUGAG-UAACCUACCUCGACCUCCGCAUCGGAGUCCUCCGACAAGUUCUCGACCUCGGCAUCGUCCUCAAUGGUGCCUAUCAAGUCUAGGUUCUUGGUUGCGCC ..(((-((((..(((.(((-..............)))))).(((((....)))))((((.(((.(((...((((((((.....))))))))......))).)))..)))).))))))) ( -34.94) >DroYak_CAF1 5761 116 - 1 UCGGA-UUAAAAGUGUGAG-UAACCUACCUCGACCUCCGCAUCGGAGUCCUCCGACAAGUUCUCGACCUCGGCAUCGUCCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACC (((.(-(.....)).)))(-(((((.((((.(((.......(((((....)))))...............(((((((((...)))))))))......))).))))....))))))... ( -37.20) >DroAna_CAF1 2684 112 - 1 -----UGUAACAGC-UGAUUUCUCUCACCUCCACUUCGGCAUCGGAGUCCUCUGAAAGGUUCUCUACCUCUGCAUCAUCCUCGAUGGUGCCAAUCAAGUCCAGGUUCUUGGUUGCACC -----.......((-(((.................))))).(((((....))))).((((.....))))..((((((((...))))))))((((((((........)))))))).... ( -29.13) >consensus UCGGUUUUAAAAGUGUGAG_UAACCUACCUCGACCUCCGCAUCGGAGUCCUCCGACAAGUUCUCCACCUCGGCAUCGUCCUCGAUGGUGCCUAUCAAGUCCAGGUUCUUGGUUGCACC ............((((((.....((.((((.(((.......(((((....)))))...............(((((((((...)))))))))......))).))))....)))))))). (-27.42 = -27.66 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:23 2006