| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,662,514 – 3,662,617 |
| Length | 103 |
| Max. P | 0.657615 |

| Location | 3,662,514 – 3,662,617 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
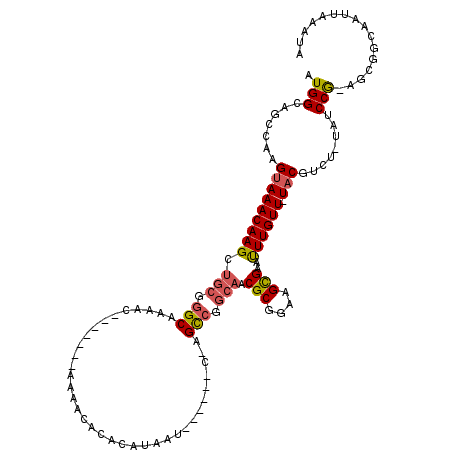
>2R_DroMel_CAF1 3662514 103 - 20766785 AUGGCAGCCGAGUAAACAAGCUGCGGGCAAAAC-------AAAACAUACAUAAG------C-AGCCGGCAACGCGGAAGCGAAUUUGUU-UACGUCU-UAUCCA-AACGGCAAUUAAAUA ......((((.(((((((((.(((.(((.....-------..............------.-.))).))).(((....)))..))))))-)))(...-...)..-..))))......... ( -29.10) >DroSec_CAF1 10987 103 - 1 AUGGCAGCCAAGUAAACAAGCUGCGGGCAAAAC-------AAAACACACAUAAU------C-AGCCGGCAACGCGGAAGCGAAUUUGUU-UACGUCU-UAGCCG-AGCGGCAAUUAAAUA ......(((..(((((((((.(((.(((.....-------..............------.-.))).))).(((....)))..))))))-)))(.((-(....)-))))))......... ( -29.60) >DroSim_CAF1 14844 103 - 1 AUGGCAGCCAAGUAAACAAGCUGCGGGCAAAAC-------AAAACACACAUAAU------C-AGCCGGCAACGCGGAAGCGAAUUUGUU-UACGUCU-UAUCCG-AGCGGCAAUUAAAUA ......(((..(((((((((.(((.(((.....-------..............------.-.))).))).(((....)))..))))))-)))(.((-(....)-))))))......... ( -29.60) >DroEre_CAF1 10423 101 - 1 AUGGCAGCCAAGUAAACAAGCUGCGGGCAAAAC-------AAAAC--ACACCAG------C-AGCCGGCAACGCGGAAGCGAAUUUGUU-UACGUCU-UAUCCG-AGUCUCAAUUAGAUA .(((.......(((((((((((((((.......-------.....--...)).)------)-)))......(((....)))...)))))-)))....-...)))-.((((.....)))). ( -27.10) >DroYak_CAF1 11102 108 - 1 AUGGCAGCCAAGUAAACAAGCAGCGGGCAAAACAAAAAACAAAAC--ACACCAU------C-AGCCGGCAACGCGGAAGCGAACUUGUU-UACGUCU-UAUCCG-AGCGGCAAUUAAAUA ......(((..(((((((((..((.(((.................--.......------.-.))).))..(((....)))..))))))-)))(.((-(....)-))))))......... ( -30.87) >DroAna_CAF1 8667 110 - 1 AUGGGAGGCAAGUAAACAAGCU---GGCAAAAA-------AAAUAACAACUAUAUAGUGGCGGGUGGGCAACGCGGAAGUGACUUUGUUUUUCGUCUUUAUCCAAAGCAGUAAUUAGAAA .((((((((....(((((((((---.((.....-------........(((....))).....)).)))..(((....)))...))))))...)))))...)))................ ( -19.87) >consensus AUGGCAGCCAAGUAAACAAGCUGCGGGCAAAAC_______AAAACACACAUAAU______C_AGCCGGCAACGCGGAAGCGAAUUUGUU_UACGUCU_UAUCCG_AGCGGCAAUUAAAUA .(((.......(((((((((.(((.(((...................................))).))).(((....)))..)))))).)))........)))................ (-16.75 = -16.94 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:14 2006