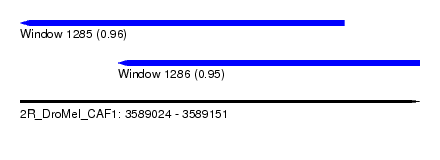
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,589,024 – 3,589,151 |
| Length | 127 |
| Max. P | 0.955071 |

| Location | 3,589,024 – 3,589,127 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
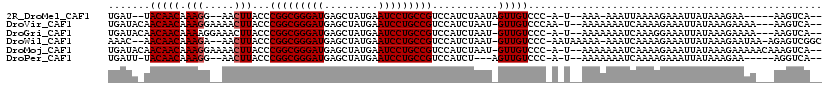
>2R_DroMel_CAF1 3589024 103 - 20766785 UGAU--UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAUAGUUGUCCC-A-U--AAA-AAAUUAAAAGAAAUUAUAAAGAA-----AAGUCA-- .(((--((((((...((--......))(((((((((.........)))))))))...........)))))...-.-.--...-...(((.((....)).)))....-----.)))).-- ( -21.30) >DroVir_CAF1 1730 110 - 1 UGAUACAACAACAAAGGAAAACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCCAA-U--AAAAAAAUCAAAAGAAAUUAUAAAGAAAA---AAGUCA-- ((..((((((...(((.....)))...(((((((((.........))))))))).........)-)))))..)).-.--.............................---......-- ( -22.00) >DroGri_CAF1 1568 109 - 1 UGAUACAACAACAAAAGGAAACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCC-A-U--AAAAAAAUCAAAGGAAAUUAUAAAGAAAA---AAGUCA-- ((..((((((....(((....)))...(((((((((.........))))))))).........)-)))))..)-)-.--.............................---......-- ( -24.80) >DroWil_CAF1 1844 111 - 1 AAAC--AACAACAAAGA--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCC-AAUAAAAA-AAAUCAAAAGAAAUUAUAAAGAAUAA-AGAGUCGGC ....--.((((((.(((--........(((((((((.........)))))))))....)))..)-)))))...-........-..........................-......... ( -19.00) >DroMoj_CAF1 2176 112 - 1 UGAUACAACAACAAAGGAAAACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCC-A-U--AAAAAAAUCAAAAGAAAUUAUAAAGAAAAACAAAGUCA-- ((..((((((...(((.....)))...(((((((((.........))))))))).........)-)))))..)-)-.--......................................-- ( -21.80) >DroPer_CAF1 1417 102 - 1 UGAUU-UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCU---AGUUGUCCC-A-U--AAAAAAAUCAAAAGAAAUUAUAAAGAA-----AGGUCA-- (((((-((((((...((--......))(((((((((.........))))))))).......---.)))))...-.-.--....)))))).................-----......-- ( -21.90) >consensus UGAUA_AACAACAAAGG__AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU_GUUGUCCC_A_U__AAAAAAAUCAAAAGAAAUUAUAAAGAAAA___AAGUCA__ .......(((((.(((.....)))...(((((((((.........)))))))))...........)))))................................................. (-18.90 = -19.07 + 0.17)
| Location | 3,589,055 – 3,589,151 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
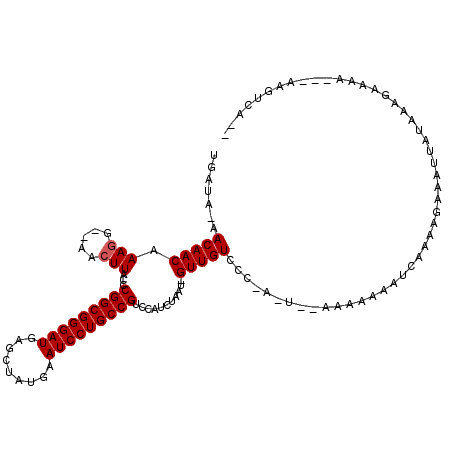
>2R_DroMel_CAF1 3589055 96 - 20766785 ----CCA-------AGAAUUA-AGCAA-CU-UUACACGUGAU--UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAUAGUUGUCCCA-U--A ----...-------.......-.((((-((-......((...--....))...((--(.......(((((((((.........)))))))))))).......))))))....-.--. ( -21.51) >DroPse_CAF1 1469 95 - 1 ----CCA-------GAAAUUA-AGCAA-CUUUUACACAUGAUU-UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCU---AGUUGUCCCA-U--A ----...-------.......-.((((-((.....((......-.........((--.......))((((((((.........))))))))))......---))))))....-.--. ( -20.30) >DroGri_CAF1 1602 85 - 1 ----------------------------CUUAUUCACUUGAUACAACAACAAAAGGAAACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCCA-U--A ----------------------------..........((..((((((....(((....)))...(((((((((.........))))))))).........)-)))))..))-.--. ( -25.00) >DroWil_CAF1 1881 99 - 1 AGAUCCA-------AAAACUA-AACAAACU-G----CAAAAC--AACAACAAAGA--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAU-GUUGUCCCAAUAAA .......-------.......-........-.----......--.((((((.(((--........(((((((((.........)))))))))....)))..)-)))))......... ( -19.00) >DroAna_CAF1 1475 105 - 1 ----CCACCAAAAAAAAAUUAUAGCAA-CUCUUACAUGUGAU--UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAUUGUUGUCCCA-U--A ----...................((((-(..(((.(((.(((--.........((--.......))((((((((.........)))))))))))))).)))..)))))....-.--. ( -23.80) >DroPer_CAF1 1449 95 - 1 ----CCA-------GAAAUUA-AGCAA-CUUUUACACAUGAUU-UACAACAAAGG--AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCU---AGUUGUCCCA-U--A ----...-------.......-.((((-((.....((......-.........((--.......))((((((((.........))))))))))......---))))))....-.--. ( -20.30) >consensus ____CCA_______AAAAUUA_AGCAA_CUUUUACACAUGAU__UACAACAAAGG__AACUUACCCGGCGGGAUGAGCUAUGAAUCCUGCCGUCCAUCUAAUAGUUGUCCCA_U__A .............................................(((((.(((.....)))...(((((((((.........)))))))))...........)))))......... (-18.90 = -19.07 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:50 2006