
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,579,944 – 3,580,054 |
| Length | 110 |
| Max. P | 0.624599 |

| Location | 3,579,944 – 3,580,054 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3579944 110 - 20766785 GCUCUUAUGUAAACCAAGGGACCCAGUAA-----UUGACGACGA--CCGCCGCUCCAGACAGCGGCC-ACUAUUGAAUUUUCAACCGAGUUUUUCGCUGACAAGCCCCCGGGAGACAU-- ......((((...((..(((...(((..(-----..(((..((.--..((((((......)))))).-....((((....)))).)).)))..)..))).....)))..))...))))-- ( -28.00) >DroPse_CAF1 2127 117 - 1 UCUCUUAUGUAAACUUAUGGCCACAGUAAUUAUAUUGACGGCGACACUGCCGCUCCAGACAGCGGCG-AGUAUUGAAUUUUCAACAAAGUUUUACGCCAUCAAAAUAUGGACAAACAU-- ......((((.......((....))((...(((.((((.((((..(((((((((......)))))).-))).((((....))))..........)))).)))).)))...))..))))-- ( -26.90) >DroEre_CAF1 6980 110 - 1 GCUCUUAUGUAAACCAAGGGACCCACUAA-----UUGACGGCGA--CCGCCGCUCCAGACAGCGGCC-ACUAUUGAAUUUUCAAGCGAGUUUUUCGCUGACAAGGCCCCGAGAGGCAA-- .(((((.((.....)).(((.((......-----(((.((((((--..((((((......)))))).-(((.((((....))))...)))...)))))).))))).))))))))....-- ( -32.20) >DroYak_CAF1 2734 110 - 1 GCUCUUAUGUAAACCAUGGGACCCAGUAA-----UUGACGGCGA--CCGCCGCUCCAGACAGCGGCC-ACUAUUGAAUUUUCAAGGGAAUUUUACGCUGACAAGGCCCCGGAAAAUAA-- ..(((((((.....)))))))((......-----(((.(((((.--..((((((......)))))).-.((.((((....)))).)).......))))).)))......)).......-- ( -28.30) >DroAna_CAF1 1266 116 - 1 ACUCUUAUGUAAAUCAAGGACCCUAGCAAUUAUAUUGACGGCGACACUGCCGCUCCAGACAGCGGCAGACUAUUGAAUUUUCAAUAGAUUUUUCCGCUGUCAAA----CGAAUGACGUCC .................((((.......(((...(((((((((...((((((((......)))))))).(((((((....))))))).......))))))))).----..)))...)))) ( -38.70) >DroPer_CAF1 2152 117 - 1 UCUCUUAUGUAAACUUAUGGCCACAGUAAUUAUAUUGACGGCGACACUGCCGCUCCAGACAGCGGCG-AGUAUUGAAUUUUCAACAAAGUUUUACGCCAUAAAAAUAUGGACAAACAU-- ........((((((((.(((((.(((((....)))))..)))...(((((((((......)))))).-))).............))))).)))))((((((....))))).)......-- ( -27.60) >consensus GCUCUUAUGUAAACCAAGGGACCCAGUAA_____UUGACGGCGA__CCGCCGCUCCAGACAGCGGCC_ACUAUUGAAUUUUCAACAGAGUUUUACGCUGACAAAACCCCGAAAAACAU__ ..................................(((.(((((.....((((((......))))))......((((....))))..........))))).)))................. (-18.09 = -18.53 + 0.45)

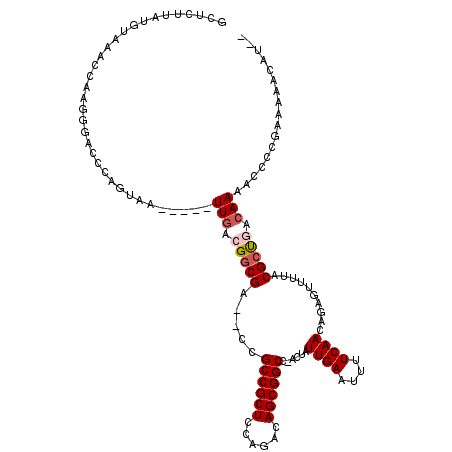
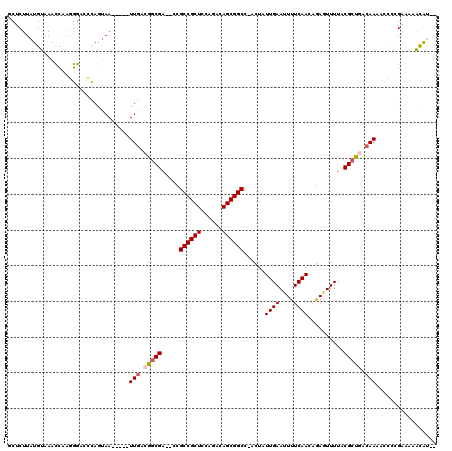
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:48 2006