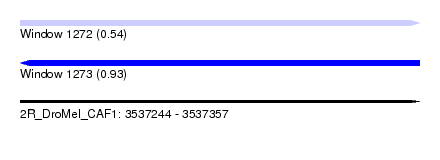
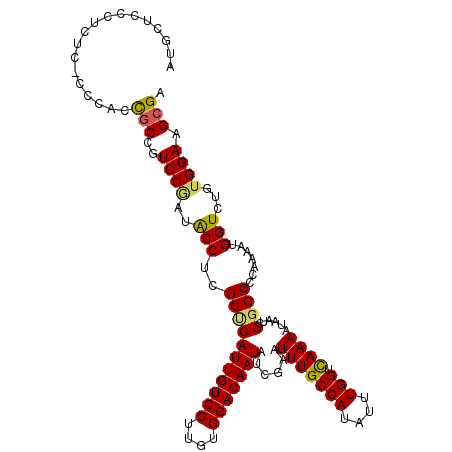
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,537,244 – 3,537,357 |
| Length | 113 |
| Max. P | 0.930307 |

| Location | 3,537,244 – 3,537,357 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -24.52 |
| Energy contribution | -26.02 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3537244 113 + 20766785 UCGCUUCCACAGACCAUUUUGGGCACGAUUAUUUUAACCAAAUAUGGCAAAUUCGAUAUUGUCCACAAGGACAAUGAGCGUGGUAUUGGAUGGCGGAGGGAGAGAGGGUGCAU ((.(((((.(.(.((((((..(.((((..........(((....))).........((((((((....))))))))..)))).)...))))))).).))))).))........ ( -31.10) >DroSec_CAF1 9677 112 + 1 UCGCUUCCACAGACCAUUUUGGGCCCGAUUAUUUUGACCAAAUAUGGCAAAUUCGAUAUUGUCCACAAGGACAAUGAGCGAGGUAUCGGACGGCGGUGGG-GAGAGGGAGCAU ..((((((.(...((((..(.(..(((((...((((.(((....)))))))((((.((((((((....))))))))..))))..))))).).)..)))).-..).)))))).. ( -39.60) >DroSim_CAF1 7334 113 + 1 UCGCUUCCACAGACCAUUUUGGGCCCGAUUAUUUUGACCAAAUAUGGCAAAUUCGAUAUUGUCCACAAGGACAAUGAGCGAGGUAUCGGACGGCAGUGGGAGAGAGGGAGCAU ..((((((.(...(((((.(.(..(((((...((((.(((....)))))))((((.((((((((....))))))))..))))..))))).).).)))))....).)))))).. ( -38.50) >DroEre_CAF1 4855 105 + 1 UCGCUUCCACAGACCAUU-UGGGCCCGAUUAUUUUGACCAAAUAUGGCAAAUUCGAUAUUGUCCACAAGGACAAUGGGCGCGGC---GGAGGGAGGCUGU-GAGGG---GGAU ((.((..(((((.((..(-((.((((((....((((.(((....))))))).))...(((((((....))))))))))).))).---...))....))))-)..))---.)). ( -38.00) >consensus UCGCUUCCACAGACCAUUUUGGGCCCGAUUAUUUUGACCAAAUAUGGCAAAUUCGAUAUUGUCCACAAGGACAAUGAGCGAGGUAUCGGACGGCGGUGGG_GAGAGGGAGCAU ((((((((...(.(((((((((...(((.....))).))))).)))))...((((.((((((((....))))))))..)))).....))).)))))................. (-24.52 = -26.02 + 1.50)

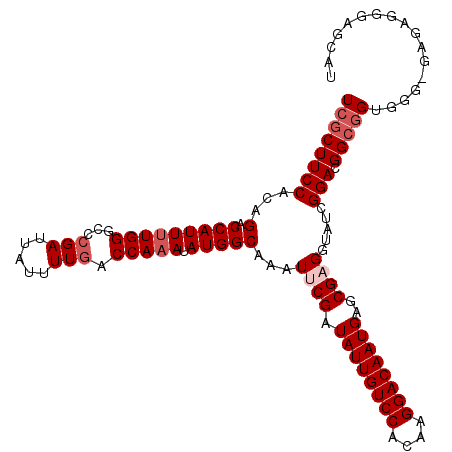
| Location | 3,537,244 – 3,537,357 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -25.61 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3537244 113 - 20766785 AUGCACCCUCUCUCCCUCCGCCAUCCAAUACCACGCUCAUUGUCCUUGUGGACAAUAUCGAAUUUGCCAUAUUUGGUUAAAAUAAUCGUGCCCAAAAUGGUCUGUGGAAGCGA ..................(((..((((..((((.....(((((((....)))))))..(((((((((((....))))..))))..))).........))))...)))).))). ( -25.90) >DroSec_CAF1 9677 112 - 1 AUGCUCCCUCUC-CCCACCGCCGUCCGAUACCUCGCUCAUUGUCCUUGUGGACAAUAUCGAAUUUGCCAUAUUUGGUCAAAAUAAUCGGGCCCAAAAUGGUCUGUGGAAGCGA .((((.((.(..-.(((.....(((((((...(((...(((((((....)))))))..))).(((((((....))).))))...)))))))......)))...).)).)))). ( -35.00) >DroSim_CAF1 7334 113 - 1 AUGCUCCCUCUCUCCCACUGCCGUCCGAUACCUCGCUCAUUGUCCUUGUGGACAAUAUCGAAUUUGCCAUAUUUGGUCAAAAUAAUCGGGCCCAAAAUGGUCUGUGGAAGCGA .((((.((.(...((.......(((((((...(((...(((((((....)))))))..))).(((((((....))).))))...))))))).......))...).)).)))). ( -34.54) >DroEre_CAF1 4855 105 - 1 AUCC---CCCUC-ACAGCCUCCCUCC---GCCGCGCCCAUUGUCCUUGUGGACAAUAUCGAAUUUGCCAUAUUUGGUCAAAAUAAUCGGGCCCA-AAUGGUCUGUGGAAGCGA ....---.....-...((.((((...---((((.(((((((((((....)))))))......(((((((....))).))))......))))...-..))))..).))).)).. ( -30.90) >consensus AUGCUCCCUCUC_CCCACCGCCGUCCGAUACCUCGCUCAUUGUCCUUGUGGACAAUAUCGAAUUUGCCAUAUUUGGUCAAAAUAAUCGGGCCCAAAAUGGUCUGUGGAAGCGA ..................(((..((((..(((..(((((((((((....)))))))......(((((((....))).))))......)))).......)))...)))).))). (-25.61 = -25.55 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:38 2006