
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,325,374 – 1,325,677 |
| Length | 303 |
| Max. P | 0.999736 |

| Location | 1,325,374 – 1,325,484 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -21.58 |
| Energy contribution | -23.70 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325374 110 + 20766785 CCUCCAGAACUGCAGAUCUCCAGACCGCUUCCUGGGUCCUUAUGUACCUUAUACA----------CGCCAUGACCAUGCAUGGCAACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAG .((((((((..((((.((....)).((((((.((((..(...((((.....))))----------.((((((......))))))...)..)))).))))))....)))))))))).)).. ( -37.90) >DroSec_CAF1 43887 94 + 1 CCUCCAGAACUCC--------AGACCGAUCCCUG--------UGUACCUUAUACA----------CGCCAUCACCAUGCAUGGCGACAAGCCCAAGGAGCGCCAACUGUUUUUGGCAGAG .(((.....((((--------............(--------((((.....))))----------)((((((.....).)))))...........)))).(((((......))))).))) ( -24.40) >DroSim_CAF1 22988 102 + 1 CCUCCAGAACUCCAGACCUCCAGACCGCUCCCUG--------UGUACCUUAUACA----------CGCCAUCACCAUGCAUGGCGACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAG .........(((((((....(((..((((((..(--------((((.....))))----------)((((((.....).)))))...........))))))....)))..))))).)).. ( -27.80) >DroEre_CAF1 25194 103 + 1 UCUUCA----------------GACGGCUCCCCAGGUUC-CAUGUACCAUGUACAGCUAUGGCGAAGCUAUGGCUGUGCAUGGCGACGAGCCCAGGGAGCGACAACUGCUUUUGGCAGAG ......----------------....((((((..(((((-...((.(((((((((((((((((...)))))))))))))))))..)))))))..)))))).....((((.....)))).. ( -52.80) >DroYak_CAF1 11261 94 + 1 UCUUCC----------------ACCCGCUCCCUGGGUUCUUAUGUACCUUAUACA----------CGCCAUGUCCAUGCAUGCCGACGAGCCCAGGGAGCGACAACUGUUUUUGGCAGAG ......----------------...((((((((((((((...((((.....))))----------((.(((((....))))).))..))))))))))))))....((((.....)))).. ( -41.00) >consensus CCUCCAGAACU_C________AGACCGCUCCCUGGGU_C__AUGUACCUUAUACA__________CGCCAUGACCAUGCAUGGCGACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAG .........................((((((.(((((.....((((.....))))...........((((((......)))))).....))))).))))))....((((.....)))).. (-21.58 = -23.70 + 2.12)

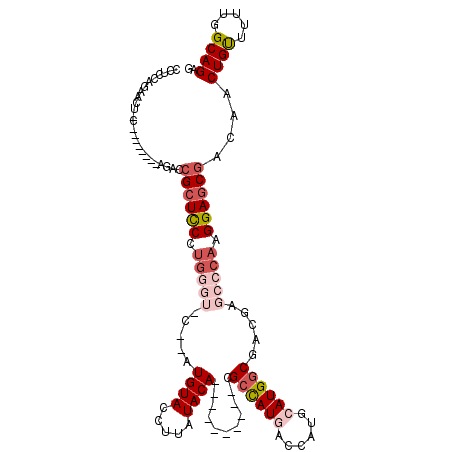
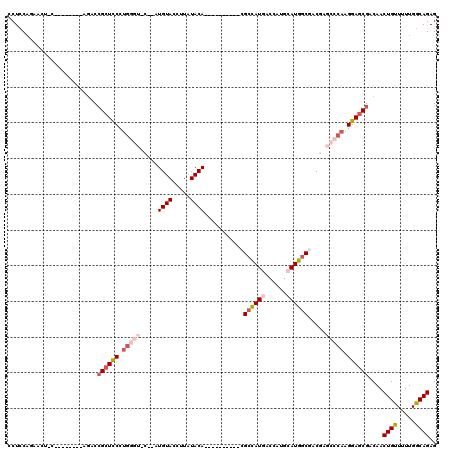
| Location | 1,325,374 – 1,325,484 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -22.28 |
| Energy contribution | -24.16 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325374 110 - 20766785 CUCUGCCAAAAACAGUUGUCGCUCCUUGGGCUCGUUGCCAUGCAUGGUCAUGGCG----------UGUAUAAGGUACAUAAGGACCCAGGAAGCGGUCUGGAGAUCUGCAGUUCUGGAGG ..((((......(((...(((((.((((((..(...((((((......))))))(----------(((((...))))))..)..)))))).))))).))).......))))......... ( -37.62) >DroSec_CAF1 43887 94 - 1 CUCUGCCAAAAACAGUUGGCGCUCCUUGGGCUUGUCGCCAUGCAUGGUGAUGGCG----------UGUAUAAGGUACA--------CAGGGAUCGGUCU--------GGAGUUCUGGAGG (((((((((......)))))(((((..(((((.((((((((........)))))(----------(((((...)))))--------)...))).)))))--------)))))...)))). ( -36.30) >DroSim_CAF1 22988 102 - 1 CUCUGCCAAAAACAGUUGUCGCUCCUUGGGCUCGUCGCCAUGCAUGGUGAUGGCG----------UGUAUAAGGUACA--------CAGGGAGCGGUCUGGAGGUCUGGAGUUCUGGAGG (((((((.....(((...(((((((((..((.((((((((....))))))))))(----------(((((...)))))--------)))))))))).)))..)))..))))......... ( -40.60) >DroEre_CAF1 25194 103 - 1 CUCUGCCAAAAGCAGUUGUCGCUCCCUGGGCUCGUCGCCAUGCACAGCCAUAGCUUCGCCAUAGCUGUACAUGGUACAUG-GAACCUGGGGAGCCGUC----------------UGAAGA ..((((.....)))).....((((((..((.((((.((((((.(((((.((.((...)).)).))))).))))))))...-)).))..))))))....----------------...... ( -40.70) >DroYak_CAF1 11261 94 - 1 CUCUGCCAAAAACAGUUGUCGCUCCCUGGGCUCGUCGGCAUGCAUGGACAUGGCG----------UGUAUAAGGUACAUAAGAACCCAGGGAGCGGGU----------------GGAAGA .((..((....((....))(((((((((((.((...(.((((......)))).)(----------(((((...))))))..)).))))))))))))).----------------.))... ( -41.40) >consensus CUCUGCCAAAAACAGUUGUCGCUCCUUGGGCUCGUCGCCAUGCAUGGUCAUGGCG__________UGUAUAAGGUACAU__G_ACCCAGGGAGCGGUCU________G_AGUUCUGGAGG ((((..(((......))).(((((((((((......((((((......))))))...........(((((...)))))......)))))))))))....................)))). (-22.28 = -24.16 + 1.88)

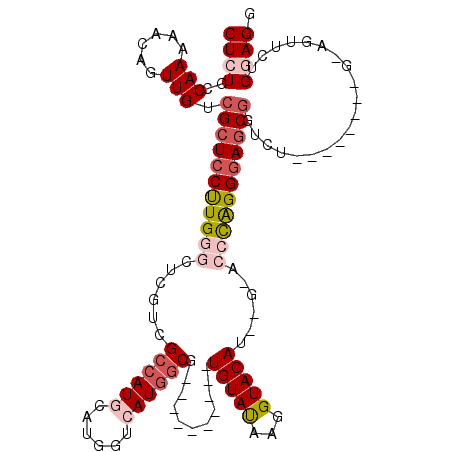
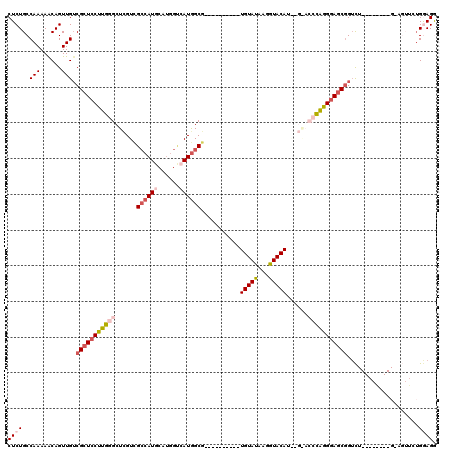
| Location | 1,325,414 – 1,325,524 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325414 110 + 20766785 UAUGUACCUUAUACA----------CGCCAUGACCAUGCAUGGCAACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAGCGCUUUUCUAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA ..((((.....))))----------.((((((......)))))).....((...(((((((....((((.....))))..)))))))..........(((((......)))))))..... ( -24.50) >DroSec_CAF1 43913 108 + 1 --UGUACCUUAUACA----------CGCCAUCACCAUGCAUGGCGACAAGCCCAAGGAGCGCCAACUGUUUUUGGCAGAGCGCUUUUCUAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA --((((.....))))----------(((((((.....).))))))....((...((((((((...((((.....)))).))))))))..........(((((......)))))))..... ( -25.60) >DroSim_CAF1 23022 108 + 1 --UGUACCUUAUACA----------CGCCAUCACCAUGCAUGGCGACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAGCGCUUUUCUAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA --((((.....))))----------(((((((.....).))))))....((...(((((((....((((.....))))..)))))))..........(((((......)))))))..... ( -23.00) >DroEre_CAF1 25217 120 + 1 CAUGUACCAUGUACAGCUAUGGCGAAGCUAUGGCUGUGCAUGGCGACGAGCCCAGGGAGCGACAACUGCUUUUGGCAGAGCGCUUUUCCAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA ..(((.(((((((((((((((((...)))))))))))))))))..))).((...(((((((....((((.....))))..)))))..))........(((((......)))))))..... ( -45.90) >DroYak_CAF1 11285 110 + 1 UAUGUACCUUAUACA----------CGCCAUGUCCAUGCAUGCCGACGAGCCCAGGGAGCGACAACUGUUUUUGGCAGAGCGUUUUCCUAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA ..((((.....))))----------((.(((((....))))).))....((..((((((((....((((.....))))..)).))))))........(((((......)))))))..... ( -20.60) >consensus _AUGUACCUUAUACA__________CGCCAUGACCAUGCAUGGCGACGAGCCCAAGGAGCGACAACUGUUUUUGGCAGAGCGCUUUUCUAUUAAAAUUUUCAUUAAAUUGAAAGCAAUUA ..((((.....))))...........((((((......)))))).....((...(((((((....((((.....))))..)))))))..........(((((......)))))))..... (-21.72 = -21.44 + -0.28)

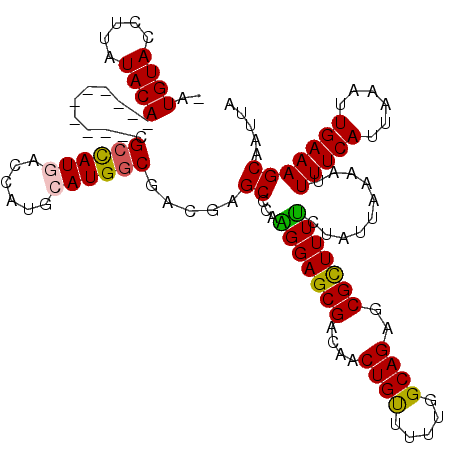
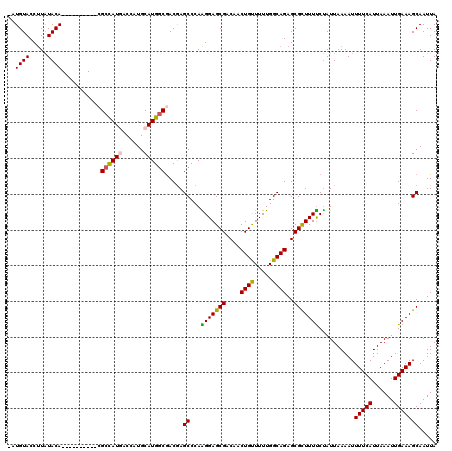
| Location | 1,325,484 – 1,325,602 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.18 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.96 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325484 118 - 20766785 ACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGGUCCCAGCCCGCCC-C-CCACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUAGAAAAGCG ......((((.(((((((((((...((((....))))..)))))..))))))))))(((....))).....-.-...........((((((.(((.(((....)))..))).).))))). ( -33.00) >DroSec_CAF1 43981 113 - 1 ACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCUC-----CCGCCC-C-ACACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUAGAAAAGCG ......((((.(((((((((((...((((....))))..)))))..))))))))))....-----......-.-...........((((((.(((.(((....)))..))).).))))). ( -31.80) >DroSim_CAF1 23090 113 - 1 ACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCAC-----CCGCCC-C-ACACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUAGAAAAGCG ......((((.(((((((((((...((((....))))..)))))..))))))))))((..-----..))..-.-...........((((((.(((.(((....)))..))).).))))). ( -31.90) >DroEre_CAF1 25297 115 - 1 ACACAAACAGCAGCAUCGCUGCGACAGCUUAGAAGCUCAGCAGCCCGGGGCCUUGUGCAC-----CCUACCGCCCCACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUGGAAAAGCG .........((.(((..(((((...((((....))))..)))))..(((((...(((...-----..))).)))))........)))(((((.(..(((....)))..).)))))..)). ( -30.80) >DroYak_CAF1 11355 114 - 1 ACUCAAAAAGCAGCAUCGCUGCAACAGCUUAGGAGCUCGGCAGCCUGGGGUCUUGUGCAC-----CCUCCCGC-CCACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUAGGAAAACG ......(((((((....(((((...((((....))))..)))))..((((((....).))-----))).....-.........))))))).............................. ( -24.00) >consensus ACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCAC_____CCGCCC_C_CCACACUAAUUGCUUUCAAUUUAAUGAAAAUUUUAAUAGAAAAGCG ......((((.(((((((((((...((((....))))..)))))..)))))))))).............................((((((.(((.(((....)))..))).)).)))). (-24.32 = -24.96 + 0.64)

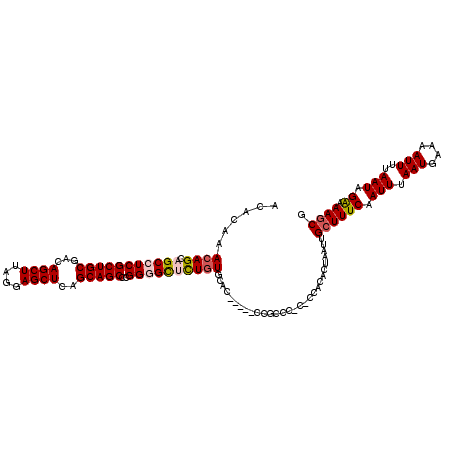
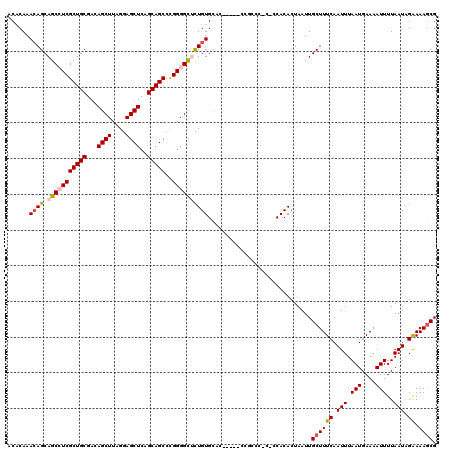
| Location | 1,325,524 – 1,325,642 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -51.94 |
| Consensus MFE | -39.71 |
| Energy contribution | -40.63 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325524 118 + 20766785 GUGUGG-G-GGGCGGGCUGGGACCACAGAGCCCCGGGCUGCUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUUAAAAGGCUGGGCAGCAGCUGCAAUUUG .(((..-(-((((...(((......))).)))))..(((((..((((....))))...)))))..(((((((((((((.(((((((....))))))).)).))))))))))))))..... ( -51.50) >DroSec_CAF1 44021 113 + 1 GUGUGU-G-GGGCGG-----GAGCACAGAGCCCCGGGCUGCUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUG ...(((-(-((((((-----((((.(((((((...)))).))).))))))..))).)))))((..(((((((((((((((((((((....)))))))))).)))))))))))))...... ( -56.00) >DroSim_CAF1 23130 113 + 1 GUGUGU-G-GGGCGG-----GUGCACAGAGCCCCGGGCUGCUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUG .(((.(-(-((((.(-----.....)...)))))).(((((..((((....))))...)))))..(((((((((((((((((((((....)))))))))).))))))))))))))..... ( -52.70) >DroEre_CAF1 25337 115 + 1 GUGUGGGGCGGUAGG-----GUGCACAAGGCCCCGGGCUGCUGAGCUUCUAAGCUGUCGCAGCGAUGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACACUGGGCAGCAGCUGCAACUCG .(((((((((((.((-----(.((.....)))))..)))))).((((....)))).)))))(((..((((((((((((((((((((....)))))))))).)))))))))))))...... ( -53.00) >DroYak_CAF1 11395 114 + 1 GUGUGG-GCGGGAGG-----GUGCACAAGACCCCAGGCUGCCGAGCUCCUAAGCUGUUGCAGCGAUGCUGCUUUUUGAGUUUUGAUUAUUGUCAAAACACUGGGCAGCAGCUGCAAUUUG .....(-((((..((-----((.......))))....))))).((((....))))((((((((..((((((((...(.((((((((....)))))))).).))))))))))))))))... ( -46.50) >consensus GUGUGG_G_GGGCGG_____GUGCACAGAGCCCCGGGCUGCUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUG .......(.((((................)))))..(((((..((((....))))...)))))(..((((((((((((((((((((....)))))))))).))))))))))..)...... (-39.71 = -40.63 + 0.92)

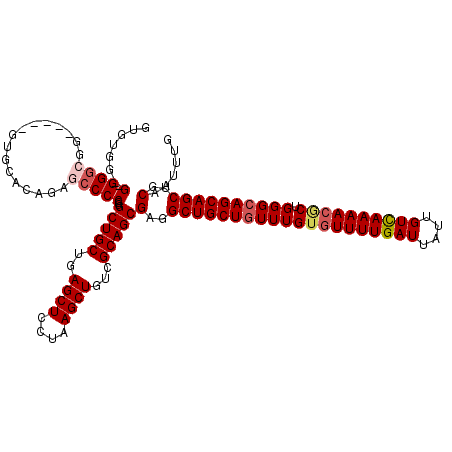
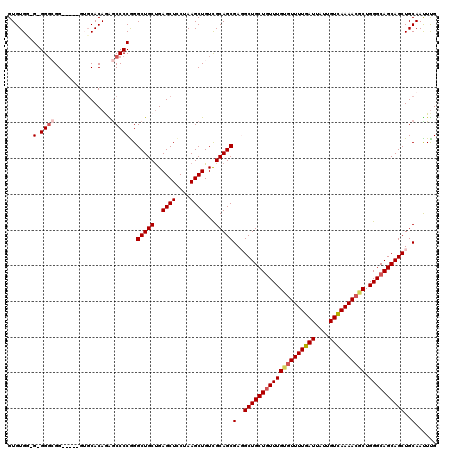
| Location | 1,325,524 – 1,325,642 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325524 118 - 20766785 CAAAUUGCAGCUGCUGCCCAGCCUUUUAACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGGUCCCAGCCCGCCC-C-CCACAC ......((.((((..(((..(........)................((((.(((((((((((...((((....))))..)))))..)))))))))))))..))))..))..-.-...... ( -34.60) >DroSec_CAF1 44021 113 - 1 CAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCUC-----CCGCCC-C-ACACAC ....(((((((.(((((...(.(((((((......))))))).).....)))))...)))))))..((...(((((.(((.((((...))))))).))))-----).))..-.-...... ( -42.60) >DroSim_CAF1 23130 113 - 1 CAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCAC-----CCGCCC-C-ACACAC .........(((((((....(.(((((((......))))))).)...)))))))...(((((...((((....))))..)))))..(((((.........-----..))))-)-...... ( -40.80) >DroEre_CAF1 25337 115 - 1 CGAGUUGCAGCUGCUGCCCAGUGUUUUGACAAUAAUCAAAACACAAACAGCAGCAUCGCUGCGACAGCUUAGAAGCUCAGCAGCCCGGGGCCUUGUGCAC-----CCUACCGCCCCACAC .........(((((((....(((((((((......)))))))))...)))))))...(((((...((((....))))..)))))..(((((...(((...-----..))).))))).... ( -45.70) >DroYak_CAF1 11395 114 - 1 CAAAUUGCAGCUGCUGCCCAGUGUUUUGACAAUAAUCAAAACUCAAAAAGCAGCAUCGCUGCAACAGCUUAGGAGCUCGGCAGCCUGGGGUCUUGUGCAC-----CCUCCCGC-CCACAC ....(((((((((((((...(.(((((((......))))))).).....))))))..)))))))..((.(((((.(((((....))))).))))).))..-----........-...... ( -38.70) >consensus CAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAGCAGCCCGGGGCUCUGUGCAC_____CCGCCC_C_CCACAC ....(((((((.(((((...(.(((((((......))))))).).....)))))...)))))))..((...(((((((.........)))))))..))...................... (-30.46 = -30.74 + 0.28)

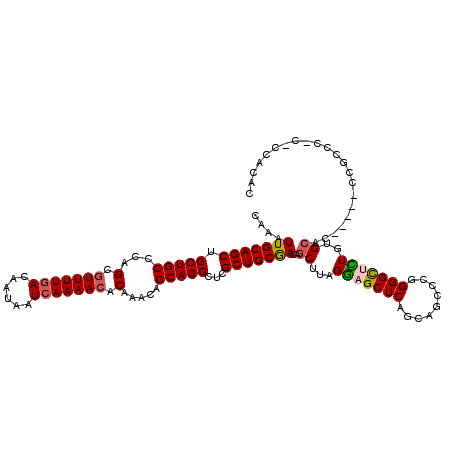
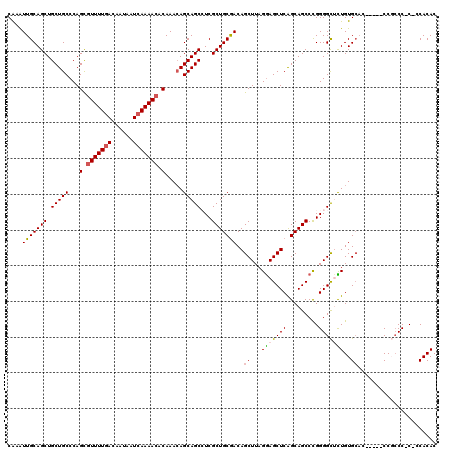
| Location | 1,325,562 – 1,325,677 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -47.28 |
| Consensus MFE | -36.74 |
| Energy contribution | -37.46 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325562 115 + 20766785 CUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUUAAAAGGCUGGGCAGCAGCUGCAAUUUGCCACCCAUAAGCGUGGCCACGUCGGCUGGGUGGGC----- ....((((((.(((((.((((((...((((((....((.(((((((....))))))).))..)))))).))))).....(((((.(....).)))))...).))))).)).))))----- ( -48.50) >DroSec_CAF1 44054 115 + 1 CUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUGCCACCCACAAACGUGGCCACGUCGCCUGGGUGGGC----- ...((((....))))...(((((...((((((....((((((((((....))))))))))..)))))).)))))......(((((((...((((....))))....)))))))..----- ( -51.40) >DroSim_CAF1 23163 97 + 1 CUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUGCCACCCGCAAACGUGG------------------C----- ...((((....))))...(((((...((((((....((((((((((....))))))))))..)))))).))))).....(((((........))))------------------)----- ( -41.60) >DroEre_CAF1 25372 120 + 1 CUGAGCUUCUAAGCUGUCGCAGCGAUGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACACUGGGCAGCAGCUGCAACUCGCCACCCACAAACGAGGCCACGUCGGCUGGGCGUCCCAUCU ....(((....(((((.((..((...((((((((((((((((((((....)))))))))).)))))))))).....((((...........))))))..)).))))).)))......... ( -46.40) >DroYak_CAF1 11429 120 + 1 CCGAGCUCCUAAGCUGUUGCAGCGAUGCUGCUUUUUGAGUUUUGAUUAUUGUCAAAACACUGGGCAGCAGCUGCAAUUUGCCACCCACAAUCGAGGCCACGUCGGCUGGGUGUGAUGGGU .......((((.((.((((((((..((((((((...(.((((((((....)))))))).).))))))))))))))))..))(((((.(....).((((.....)))))))))...)))). ( -48.50) >consensus CUGAGCUCCUAAGCUGUCGCAGCGAGGCUGCUGUUUGUGUUUUGAUUAUUGUCAAAACGCUGGGCAGCAGCUGCAAUUUGCCACCCACAAACGUGGCCACGUCGGCUGGGUGGGC_____ ...((((....))))...(((((...((((((....((((((((((....))))))))))..)))))).))))).....(((((........)))))....................... (-36.74 = -37.46 + 0.72)

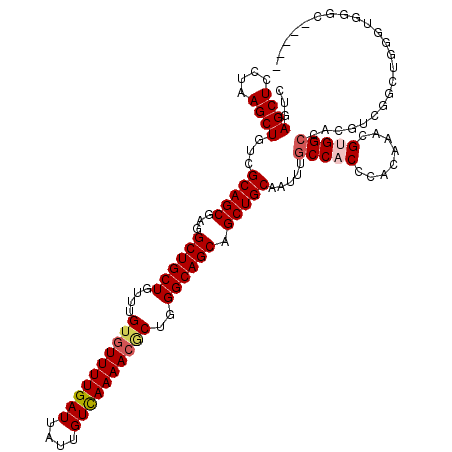
| Location | 1,325,562 – 1,325,677 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -40.81 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1325562 115 - 20766785 -----GCCCACCCAGCCGACGUGGCCACGCUUAUGGGUGGCAAAUUGCAGCUGCUGCCCAGCCUUUUAACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAG -----(((.(((((...(.(((....))))...))))))))...(((((((.(((((................................)))))...))))))).((((....))))... ( -33.75) >DroSec_CAF1 44054 115 - 1 -----GCCCACCCAGGCGACGUGGCCACGUUUGUGGGUGGCAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAG -----(((.(((((.(.(((((....)))))).))))))))...(((((((.(((((...(.(((((((......))))))).).....)))))...))))))).((((....))))... ( -44.30) >DroSim_CAF1 23163 97 - 1 -----G------------------CCACGUUUGCGGGUGGCAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAG -----(------------------((((........)))))...(((((((.(((((...(.(((((((......))))))).).....)))))...))))))).((((....))))... ( -34.60) >DroEre_CAF1 25372 120 - 1 AGAUGGGACGCCCAGCCGACGUGGCCUCGUUUGUGGGUGGCGAGUUGCAGCUGCUGCCCAGUGUUUUGACAAUAAUCAAAACACAAACAGCAGCAUCGCUGCGACAGCUUAGAAGCUCAG ...((((.((((((.(.((((......)))).)))))))((..((((((((((((((...(((((((((......))))))))).....))))))..)))))))).)).......)))). ( -51.10) >DroYak_CAF1 11429 120 - 1 ACCCAUCACACCCAGCCGACGUGGCCUCGAUUGUGGGUGGCAAAUUGCAGCUGCUGCCCAGUGUUUUGACAAUAAUCAAAACUCAAAAAGCAGCAUCGCUGCAACAGCUUAGGAGCUCGG ..((....(((((((((.....)))........))))))((...(((((((((((((...(.(((((((......))))))).).....))))))..)))))))..))...))....... ( -40.30) >consensus _____GCACACCCAGCCGACGUGGCCACGUUUGUGGGUGGCAAAUUGCAGCUGCUGCCCAGCGUUUUGACAAUAAUCAAAACACAAACAGCAGCCUCGCUGCGACAGCUUAGGAGCUCAG .......................(((((........)))))...(((((((.(((((...(.(((((((......))))))).).....)))))...))))))).((((....))))... (-29.10 = -29.94 + 0.84)

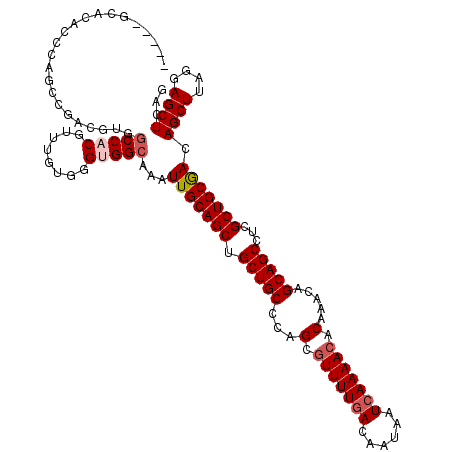
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:45 2006