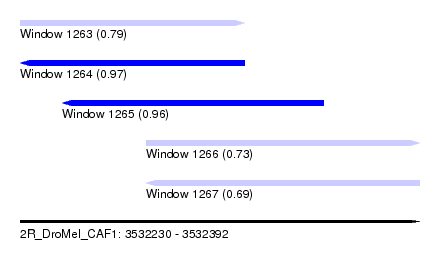
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,532,230 – 3,532,392 |
| Length | 162 |
| Max. P | 0.965920 |

| Location | 3,532,230 – 3,532,321 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 99.27 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -30.73 |
| Energy contribution | -31.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3532230 91 + 20766785 AUGGCGAGUCCAGCCCUCUCUAUGACAUUGAUGGGCAUUGACGACGCUGGUGAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCA ..((((.(((..((((((...........)).))))...)))..))))((((((((((((.....)))).((....))....)))))))). ( -32.10) >DroSec_CAF1 3590 91 + 1 AUGGCGAGUCCAGCCCUCUCUAUGACAUUGAUGGGCAUUGACGACGCUGCUGAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCA (((..(((((..((((((...........)).))))...)))((((((((((((((........))))))))...)))))).))..))).. ( -32.90) >DroSim_CAF1 1 91 + 1 AUGGCGAGUCCAGCCCUCUCUAUGACAUUGAUGGGCAUUGACGACGCUGCUGAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCA (((..(((((..((((((...........)).))))...)))((((((((((((((........))))))))...)))))).))..))).. ( -32.90) >consensus AUGGCGAGUCCAGCCCUCUCUAUGACAUUGAUGGGCAUUGACGACGCUGCUGAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCA (((..(((((..((((((...........)).))))...)))((((((((((((((........))))))))...)))))).))..))).. (-30.73 = -31.07 + 0.33)

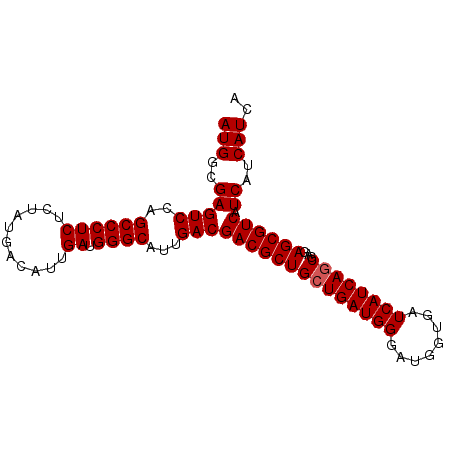
| Location | 3,532,230 – 3,532,321 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 99.27 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -30.93 |
| Energy contribution | -31.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3532230 91 - 20766785 UGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUCACCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAGAGGGCUGGACUCGCCAU ......((((((((((...(.(((((..........))))).)))))))))))..((((.((.........))..))))(((.....))). ( -26.50) >DroSec_CAF1 3590 91 - 1 UGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUCAGCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAGAGGGCUGGACUCGCCAU ......((((((((((...(((((((..........)))))))))))))))))..((((.((.........))..))))(((.....))). ( -33.10) >DroSim_CAF1 1 91 - 1 UGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUCAGCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAGAGGGCUGGACUCGCCAU ......((((((((((...(((((((..........)))))))))))))))))..((((.((.........))..))))(((.....))). ( -33.10) >consensus UGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUCAGCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAGAGGGCUGGACUCGCCAU ......((((((((((...(((((((..........)))))))))))))))))..((((.((.........))..))))(((.....))). (-30.93 = -31.27 + 0.33)

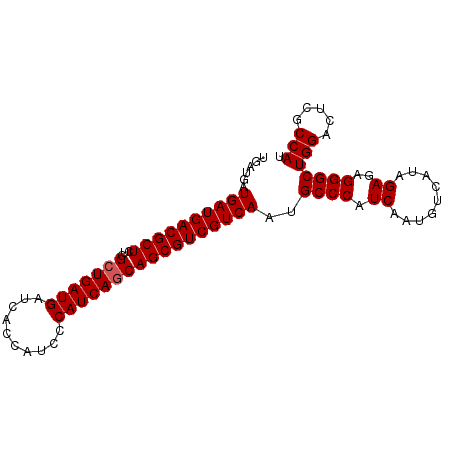
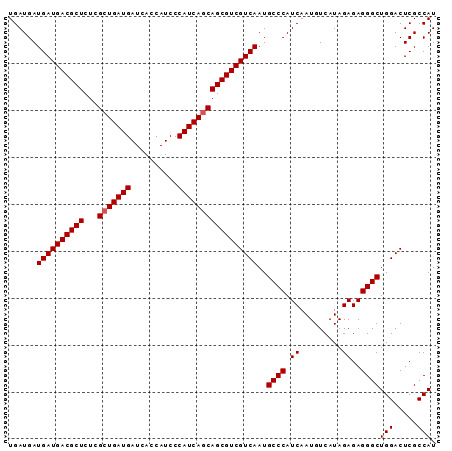
| Location | 3,532,247 – 3,532,353 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -20.28 |
| Energy contribution | -19.79 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3532247 106 - 20766785 UUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC------ACCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAG .(((((((((....(((.........)))...)))))))))(((((....(((((.((((.........)))------).))))).)))))..................... ( -26.00) >DroSec_CAF1 3607 106 - 1 UUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC------AGCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAG .(((((((((....(((.........)))...)))))))(((((((((...(((((((..........))))------))))))))))))...................)). ( -30.50) >DroSim_CAF1 18 106 - 1 UUCAUCGUCAUUCCCUGCCACAUUCCCGGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC------AGCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAG .(((((((((......(((........)))..)))))))(((((((((...(((((((..........))))------))))))))))))...................)). ( -33.70) >DroEre_CAF1 1 99 - 1 UUCAUCGUCAUUCCCUGCCACAUUCCCAGAGCUGAUGAUGAUGACACUGUUGAUGAUGAUCACCAUCCCGUGAUGACGACCAUCGUCGUCAAUGCCCAU------------- .(((((((((((.((((.........))).)..)))))))))))....(((((((((((((..((((....))))..))...)))))))))))......------------- ( -28.40) >consensus UUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC______ACCAGCGUCGUCAAUGCCCAUCAAUGUCAUAGAG .(((((((((....(((.........)))...)))))))))(((((....(((((.........................))))).)))))..................... (-20.28 = -19.79 + -0.50)

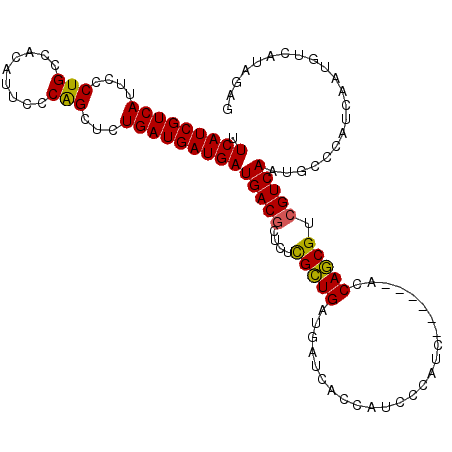
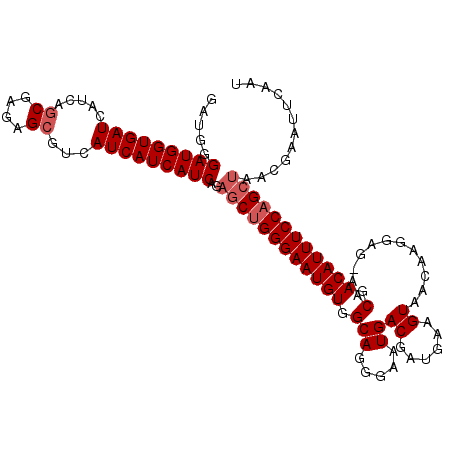
| Location | 3,532,281 – 3,532,392 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -27.95 |
| Energy contribution | -28.95 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3532281 111 + 20766785 GAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCAGAGCUGGGAAUGUGGCAGGGAAUGACGAUGAAGUAACAAGGAG-AACGACAUUUCCAGCUAACGAAUUCAAU ..((((((((((((.....((....))...)))))))))..((((((((((((.(.........((......)).........-..).)))))))))))).......))).. ( -33.85) >DroSec_CAF1 3641 111 + 1 GAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCAGAGCUGGGAAUGUGGCAGGGAAUGACGAUGAAGUAACAAGGAG-AACGACAUUUCCAGCUAACGAAUUCAAU ..((((((((((((.....((....))...)))))))))..((((((((((((.(.........((......)).........-..).)))))))))))).......))).. ( -33.85) >DroSim_CAF1 52 111 + 1 GAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCAGAGCCGGGAAUGUGGCAGGGAAUGACGAUGAAGUAACAAGGAG-AACGACAUUUCCAGCUAACGAAUUCAAU .....(((((((((.....((....))...)))))))))........((((.((((.(((((((.((................-..)).))))))).))))....))))... ( -29.67) >DroEre_CAF1 28 112 + 1 CACGGGAUGGUGAUCAUCAUCAACAGUGUCAUCAUCAUCAGCUCUGGGAAUGUGGCAGGGAAUGACGAUGAAGUAACAAGGAGGAACGACAUUUCCAGCUAACGAAUUCAAU .....(((((((((...(((.....)))..)))))))))........((((.((((.(((((((.((...................)).))))))).))))....))))... ( -26.31) >consensus GAUGGGAUGGUGAUCAUCAGCGAGAGCGUCAUCAUCAUCAGAGCUGGGAAUGUGGCAGGGAAUGACGAUGAAGUAACAAGGAG_AACGACAUUUCCAGCUAACGAAUUCAAU .....(((((((((.....((....))...)))))))))..((((((((((((.(((.....))((......))............).))))))))))))............ (-27.95 = -28.95 + 1.00)

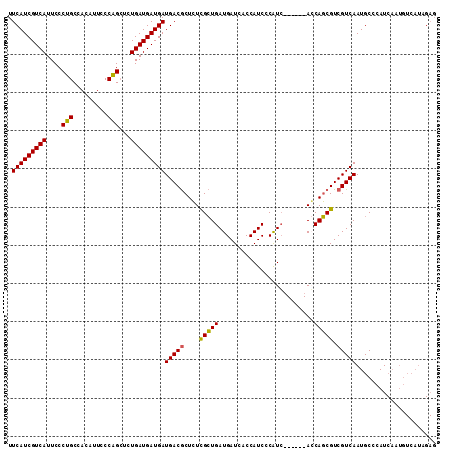
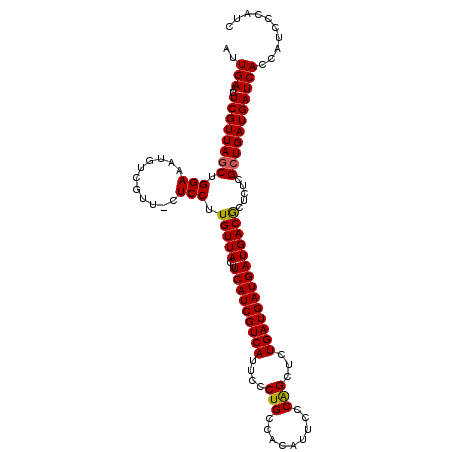
| Location | 3,532,281 – 3,532,392 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3532281 111 - 20766785 AUUGAAUUCGUUAGCUGGAAAUGUCGUU-CUCCUUGUUACUUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC ..(((..((((((((.(((..((((((.-(.....)..)).(((((((((....(((.........)))...))))))))).)))).))).))))))))))).......... ( -25.40) >DroSec_CAF1 3641 111 - 1 AUUGAAUUCGUUAGCUGGAAAUGUCGUU-CUCCUUGUUACUUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC ..(((..((((((((.(((..((((((.-(.....)..)).(((((((((....(((.........)))...))))))))).)))).))).))))))))))).......... ( -25.40) >DroSim_CAF1 52 111 - 1 AUUGAAUUCGUUAGCUGGAAAUGUCGUU-CUCCUUGUUACUUCAUCGUCAUUCCCUGCCACAUUCCCGGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC ..(((..((((((((.(((.........-.)))........(((((((((((....(((........)))...))))))))))).......))))))))))).......... ( -29.50) >DroEre_CAF1 28 112 - 1 AUUGAAUUCGUUAGCUGGAAAUGUCGUUCCUCCUUGUUACUUCAUCGUCAUUCCCUGCCACAUUCCCAGAGCUGAUGAUGAUGACACUGUUGAUGAUGAUCACCAUCCCGUG ..(((..((((((((.((((......))))....(((((..(((((((((..(.(((.........))).).))))))))))))))..))))))))...))).......... ( -25.10) >consensus AUUGAAUUCGUUAGCUGGAAAUGUCGUU_CUCCUUGUUACUUCAUCGUCAUUCCCUGCCACAUUCCCAGCUCUGAUGAUGAUGACGCUCUCGCUGAUGAUCACCAUCCCAUC ..(((..((((((((.(((...........))).(((((..(((((((((....(((.........)))...)))))))))))))).....))))))))))).......... (-22.98 = -22.85 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:32 2006