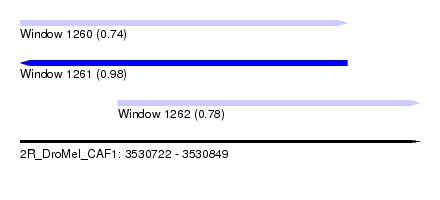
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,530,722 – 3,530,849 |
| Length | 127 |
| Max. P | 0.976438 |

| Location | 3,530,722 – 3,530,826 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -31.24 |
| Energy contribution | -34.16 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3530722 104 + 20766785 CGCCCUGUCUCCCUCCCACCGCCGUCCGGCUGCCAUUGGCAAUGGGGGCGUGGCGAGGCCCGUUUCACGCCCC--UGCUGCAGUGCAAGCGAGACAACCGUAUGGC .(((.((((((.((..(((.(((....)))........(((..((((((((((((.....)))..))))))))--)..))).)))..)).)))))).......))) ( -43.40) >DroSim_CAF1 24595 104 + 1 CGCCCUGUCUCCCUCCCACCGCCGUGCGGCUGCCAUUGGCAAAGGGGGCGUGGUAAGACCUCAUCCACGCCCC--UGCUGCAGUGCGAGCGAGACAACCGUAUGGC .(((.((((((.(((.(((.....((((((((((...))))..((((((((((...........)))))))))--)))))))))).))).)))))).......))) ( -48.80) >DroEre_CAF1 23224 103 + 1 CGCACUGUCUCCCUCCCACCAGCGUGCGGCUGCCAUUGGCAAAGG-GGCGUGGCGUGGCCUCAUCCACGCCCC--UGCAGCAGUGCGAGCGAGACAACCGUAUGGC .....((((((.(((.(((..((.(((...((((...)))).(((-(((((((.(((....))))))))))))--)))))).))).))).)))))).......... ( -47.80) >DroYak_CAF1 22339 104 + 1 CGCACUGUCUCCCUCCCACCGCCGUGUGGCUGCCAUUGGCAAAGGGGGCGUGGCGUGGCCUCAUCCACGCCCC--UGCUGCAGUGCGAGGGAGACAACCGUAUGAC .....((((((((((.(((.....((..((((((...))))..((((((((((.(((....))))))))))))--)))..))))).)))))))))).......... ( -54.70) >DroAna_CAF1 18065 95 + 1 CUCACUCUCUCUAUA----------UCGGCUGCCAUUGGCAAAGG-GGCGUGACAGGGCCAUGGCCACGCCUAGACGCUCAAGUGGAAAAGAGACAACCGUAUGAG ((((((((.(((((.----------(.(((((((...))))...(-(((((.....(((....)))))))))....))).).)))))..))))((....)).)))) ( -28.30) >consensus CGCACUGUCUCCCUCCCACCGCCGUGCGGCUGCCAUUGGCAAAGGGGGCGUGGCGAGGCCUCAUCCACGCCCC__UGCUGCAGUGCGAGCGAGACAACCGUAUGGC .....((((((.(((.(((.....((((((((((...))))...(((((((((.(((....))))))))))))...))))))))).))).)))))).......... (-31.24 = -34.16 + 2.92)

| Location | 3,530,722 – 3,530,826 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -50.12 |
| Consensus MFE | -36.72 |
| Energy contribution | -39.04 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3530722 104 - 20766785 GCCAUACGGUUGUCUCGCUUGCACUGCAGCA--GGGGCGUGAAACGGGCCUCGCCACGCCCCCAUUGCCAAUGGCAGCCGGACGGCGGUGGGAGGGAGACAGGGCG (((......(((((((.(((.((((((....--((((((((...((.....)).))))))))..(((((...))))).......)))))).))).)))))))))). ( -50.00) >DroSim_CAF1 24595 104 - 1 GCCAUACGGUUGUCUCGCUCGCACUGCAGCA--GGGGCGUGGAUGAGGUCUUACCACGCCCCCUUUGCCAAUGGCAGCCGCACGGCGGUGGGAGGGAGACAGGGCG (((......(((((((.(((.((((((.((.--(((((((((.(((....))))))))))))..(((((...)))))..))...)))))).))).)))))))))). ( -58.10) >DroEre_CAF1 23224 103 - 1 GCCAUACGGUUGUCUCGCUCGCACUGCUGCA--GGGGCGUGGAUGAGGCCACGCCACGCC-CCUUUGCCAAUGGCAGCCGCACGCUGGUGGGAGGGAGACAGUGCG (((....)))((((((.(((.((((((((((--(((((((((.((......)))))))))-)))(((((...)))))..))).)).)))).))).))))))..... ( -54.50) >DroYak_CAF1 22339 104 - 1 GUCAUACGGUUGUCUCCCUCGCACUGCAGCA--GGGGCGUGGAUGAGGCCACGCCACGCCCCCUUUGCCAAUGGCAGCCACACGGCGGUGGGAGGGAGACAGUGCG .........(((((((((((.((((((....--(((((((((.((......)))))))))))..(((((...))))).......)))))).))))))))))).... ( -57.20) >DroAna_CAF1 18065 95 - 1 CUCAUACGGUUGUCUCUUUUCCACUUGAGCGUCUAGGCGUGGCCAUGGCCCUGUCACGCC-CCUUUGCCAAUGGCAGCCGA----------UAUAGAGAGAGUGAG ((((((((((((((..........(((.(((....((((((((.........))))))))-....)))))).)))))))).----------))).)))........ ( -30.80) >consensus GCCAUACGGUUGUCUCGCUCGCACUGCAGCA__GGGGCGUGGAUGAGGCCACGCCACGCCCCCUUUGCCAAUGGCAGCCGCACGGCGGUGGGAGGGAGACAGUGCG .........(((((((.(((.((((((......(((((((((.((......)))))))))))..(((((...))))).......)))))).))).))))))).... (-36.72 = -39.04 + 2.32)

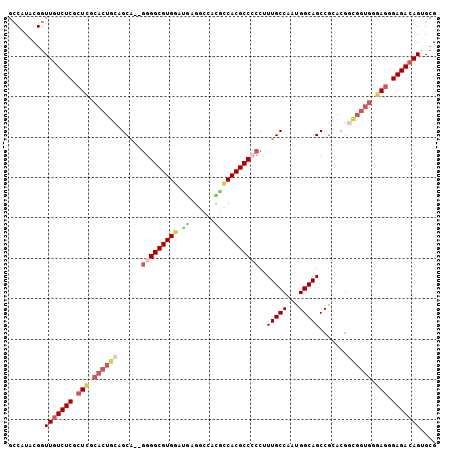
| Location | 3,530,753 – 3,530,849 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.93 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.98 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3530753 96 + 20766785 GCCAUUGGCAAUGGGGGCGUGGCGAGGCCCGUUUCACGCCCC--UGCUGCAGUGCAAGCGAGACAACCGUAU-GGCGGAGGGCUGCCAGGCAAG----UGUAU------------- (((....(((..((((((((((((.....)))..))))))))--)..))).......(((.......))).(-(((((....)))))))))...----.....------------- ( -40.60) >DroPse_CAF1 12124 111 + 1 UCCAUUGGCAAAGAGGGCGUGUCGAGAGUUGG-UCACGCCCC---ACUAUAGCAGAAAAGAGACAACCAUAUAGAUGGAACUCUUGAAGGGAAAAUACUUAAUGAACAUAC-UACU (((....((..((.(((((((((((...))))-.))))))).---.))...))....(((((....((((....))))..)))))....)))...................-.... ( -25.10) >DroSim_CAF1 24626 96 + 1 GCCAUUGGCAAAGGGGGCGUGGUAAGACCUCAUCCACGCCCC--UGCUGCAGUGCGAGCGAGACAACCGUAU-GGCGGAGGGCUGCCAGGCAAG----UGUAU------------- (((..(((((..((((((((((...........)))))))))--).((((.(((((.(........))))))-.)))).....))))))))...----.....------------- ( -42.00) >DroEre_CAF1 23255 108 + 1 GCCAUUGGCAAAGG-GGCGUGGCGUGGCCUCAUCCACGCCCC--UGCAGCAGUGCGAGCGAGACAACCGUAU-GGCCGAGCGCCGCCAGGCAAG----UGUAUGUACAUACAUACA (((..((((..(((-(((((((.(((....))))))))))))--)......((((..((.(.((....)).)-.))...)))).)))))))...----(((((((....))))))) ( -47.50) >DroYak_CAF1 22370 96 + 1 GCCAUUGGCAAAGGGGGCGUGGCGUGGCCUCAUCCACGCCCC--UGCUGCAGUGCGAGGGAGACAACCGUAU-GACGGAGGGCCGCCAGGCAAG----UGUAU------------- (((..((((...((((((((((.(((....))))))))))))--).(((..(((((..(....)...)))))-..)))......)))))))...----.....------------- ( -43.90) >DroAna_CAF1 18086 87 + 1 GCCAUUGGCAAAGG-GGCGUGACAGGGCCAUGGCCACGCCUAGACGCUCAAGUGGAAAAGAGACAACCGUAU-GAG----------UAGACUAU----GUUAU------------- .....(((((.(((-(((((.....(((....)))))))))....(((((..(((...........)))..)-)))----------)...)).)----)))).------------- ( -25.00) >consensus GCCAUUGGCAAAGGGGGCGUGGCGAGGCCUCAUCCACGCCCC__UGCUGCAGUGCAAGCGAGACAACCGUAU_GACGGAGGGCUGCCAGGCAAG____UGUAU_____________ (((...((((...(((((((((...........)))))))))..))))..................((((....))))..........)))......................... (-20.32 = -21.98 + 1.67)

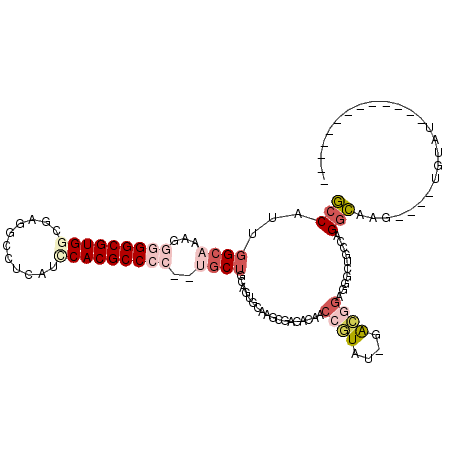
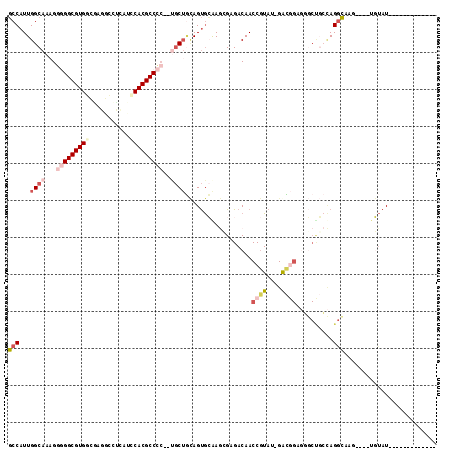
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:28 2006