
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,502,967 – 3,503,061 |
| Length | 94 |
| Max. P | 0.876352 |

| Location | 3,502,967 – 3,503,061 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -24.54 |
| Energy contribution | -24.27 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
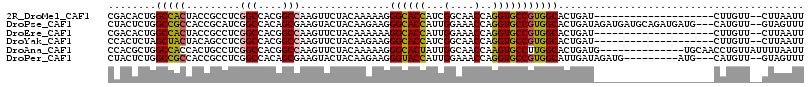
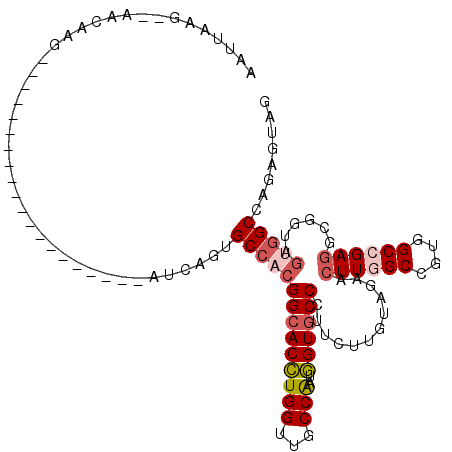
>2R_DroMel_CAF1 3502967 94 + 20766785 CGACACUGGCCACUACCGCCUCGGCCACGGCCAAGUUCUACAAAAAGGGCACCAUCGGCAACCAGGUGCCGUGGCACUGAU--------------------CUUGUU--CUUAAUU .((((...(((((.........(((....)))...............((((((...(....)..)))))))))))..)).)--------------------).....--....... ( -29.40) >DroPse_CAF1 6782 111 + 1 CUACUCUGGCCGCCACCGCAUCGGCCACAGCGAAGUACUACAAGAAGGGCACCAUUGGAAACCAGGUGCCGUGGCACUGAUAGAUGAUGCAGAUGAUG---CAUGUU--GUAGUUU ((((...((......))(((((((((((.........((......))((((((...(....)..))))))))))).(((.((.....))))).)))))---).....--))))... ( -35.50) >DroEre_CAF1 7669 94 + 1 CGACACUGGCCACUACCGCCUCGGCCACGGCCAAGUUCUACAAAAAAGGCACCAUUGGAAACCAGGUGCCGUGGCACUGAU--------------------CUUGUU--CUUAAUU .((((...(((((.........(((....)))...............((((((...(....)..)))))))))))..)).)--------------------).....--....... ( -30.10) >DroYak_CAF1 7621 94 + 1 CCACUCUAGCUACUACAGCCUCGGCCACGGCCAAGUUCUACAAGAAGGGCACCAUCGGCAACCAGGUGCCGUGGCACUGAU--------------------CUUGUU--CUUAAUU ........(((.....)))...(((....)))(((....((((((..((..(((.(((((......))))))))..))..)--------------------))))).--))).... ( -26.70) >DroAna_CAF1 7440 102 + 1 CCACGCUGGCCACCACUGCCUCGGCCACGGCCAAGUUCUACAAAAAGGGCACUAUUGGCAACCAAGUGCCUUGGCACUGAUG--------------UGCAACCUGUUAUUUUAAUU ........((((.((.((((..(((....))).............((((((((...(....)..)))))))))))).)).))--------------.))................. ( -30.00) >DroPer_CAF1 6795 102 + 1 CUACUCUGGCCGCCACCGCCUCGGCCACAGCGAAGUACUACAAGAAGGGUACCAUUGGAAACCAGGUGCCGUGGCAUUGAUAGAUG---------AUG---CAUGUU--GUAGUUU ...((.((((((.........)))))).))......(((((((.(..((((((...(....)..))))))...(((((.......)---------)))---).).))--))))).. ( -32.80) >consensus CCACUCUGGCCACCACCGCCUCGGCCACGGCCAAGUUCUACAAAAAGGGCACCAUUGGAAACCAGGUGCCGUGGCACUGAU____________________CUUGUU__CUUAAUU ........(((((.........(((....)))...............((((((...(....)..)))))))))))......................................... (-24.54 = -24.27 + -0.27)
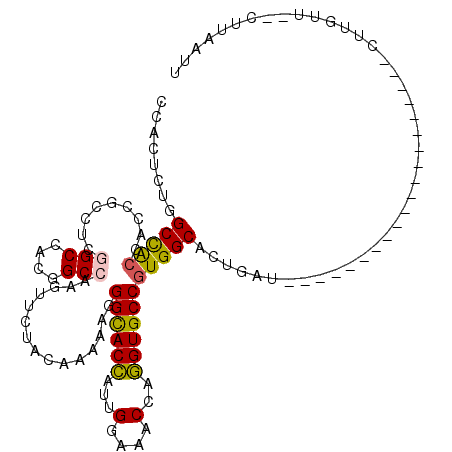
| Location | 3,502,967 – 3,503,061 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -23.51 |
| Energy contribution | -24.35 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
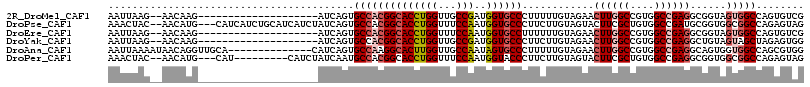
>2R_DroMel_CAF1 3502967 94 - 20766785 AAUUAAG--AACAAG--------------------AUCAGUGCCACGGCACCUGGUUGCCGAUGGUGCCCUUUUUGUAGAACUUGGCCGUGGCCGAGGCGGUAGUGGCCAGUGUCG .......--.(((((--------------------(...(..((((((((......))))).)))..)...)))))).((..(((((((..((((...))))..)))))))..)). ( -37.90) >DroPse_CAF1 6782 111 - 1 AAACUAC--AACAUG---CAUCAUCUGCAUCAUCUAUCAGUGCCACGGCACCUGGUUUCCAAUGGUGCCCUUCUUGUAGUACUUCGCUGUGGCCGAUGCGGUGGCGGCCAGAGUAG ..(((((--((.(((---((.....)))))................(((((((((...)))..))))))....)))))))((((.(((((.((((...)))).)))))..)))).. ( -37.20) >DroEre_CAF1 7669 94 - 1 AAUUAAG--AACAAG--------------------AUCAGUGCCACGGCACCUGGUUUCCAAUGGUGCCUUUUUUGUAGAACUUGGCCGUGGCCGAGGCGGUAGUGGCCAGUGUCG .......--.....(--------------------(.((.(((((((((((((((...)))..))))))............((((((....))))))......)))).)).)))). ( -34.60) >DroYak_CAF1 7621 94 - 1 AAUUAAG--AACAAG--------------------AUCAGUGCCACGGCACCUGGUUGCCGAUGGUGCCCUUCUUGUAGAACUUGGCCGUGGCCGAGGCUGUAGUAGCUAGAGUGG .......--.(((((--------------------(...(..((((((((......))))).)))..)...))))))....((((((....))))))(((.(((...))).))).. ( -36.80) >DroAna_CAF1 7440 102 - 1 AAUUAAAAUAACAGGUUGCA--------------CAUCAGUGCCAAGGCACUUGGUUGCCAAUAGUGCCCUUUUUGUAGAACUUGGCCGUGGCCGAGGCAGUGGUGGCCAGCGUGG ...........((.((((..--------------(((((.((((..(((((((((...)))..)))))).............(((((....))))))))).)))))..)))).)). ( -40.60) >DroPer_CAF1 6795 102 - 1 AAACUAC--AACAUG---CAU---------CAUCUAUCAAUGCCACGGCACCUGGUUUCCAAUGGUACCCUUCUUGUAGUACUUCGCUGUGGCCGAGGCGGUGGCGGCCAGAGUAG ..(((((--((...(---(((---------.........))))...((.((((((...)))..))).))....)))))))((((.(((((.((((...)))).)))))..)))).. ( -28.50) >consensus AAUUAAG__AACAAG____________________AUCAGUGCCACGGCACCUGGUUGCCAAUGGUGCCCUUCUUGUAGAACUUGGCCGUGGCCGAGGCGGUAGUGGCCAGAGUAG .........................................((((((((((((((...)))..))))))............((((((....))))))......)))))........ (-23.51 = -24.35 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:15 2006