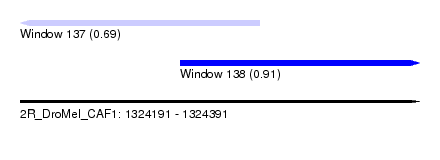
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,324,191 – 1,324,391 |
| Length | 200 |
| Max. P | 0.908142 |

| Location | 1,324,191 – 1,324,311 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -31.14 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1324191 120 - 20766785 CACGUUGAUUGGUUUUUGCCCGGAGUAAAAGCGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGCCUG ....((((((((((.(((((.(((((((..(((((.(((((((((......)))))))))..........)))))..)).....))))).)))))..))))))))))............. ( -33.60) >DroSec_CAF1 42730 120 - 1 CACGUUGAUUGGUUUUUGCCCGGAGUAGAAGCGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGCCUG ....((((((((((.(((((.(((((.((((((((.(((((((((......)))))))))..........)))))..)))....))))).)))))..))))))))))............. ( -33.70) >DroSim_CAF1 21811 120 - 1 CACGUUGAUUGGUUUUUGCCCGGAGUAGAAGCGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGCCUG ....((((((((((.(((((.(((((.((((((((.(((((((((......)))))))))..........)))))..)))....))))).)))))..))))))))))............. ( -33.70) >DroEre_CAF1 24080 120 - 1 CACGUUGAUUGGUUCUUGCCUGGAGCAAAAACGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGCCUG ....((((((((((.(((((.((((((((..((((.(((((((((......)))))))))..........))))...))))....)))).)))))..))))))))))............. ( -30.90) >DroYak_CAF1 10166 120 - 1 UACGUUGAUUGGUUUUUGCCUGGAGUGAAAUCGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGGCUG .....(((((((((.(((((.(((((.((((((((.(((((((((......)))))))))..........))))..))))....))))).)))))..)))))))))((((.....)))). ( -33.20) >consensus CACGUUGAUUGGUUUUUGCCCGGAGUAAAAGCGAUGGCUUUUGGUCACGUAAUUAAGAGCAAACUUAUUAAUCGCCGUUUGACAACUCCCGGCAGCCAGCUAAUUAAGUCAAAUUGCCUG ....((((((((((.(((((.(((((((((((....)))))))((((.(..((((((((....))).)))))..)....))))..)))).)))))..))))))))))............. (-31.14 = -30.98 + -0.16)

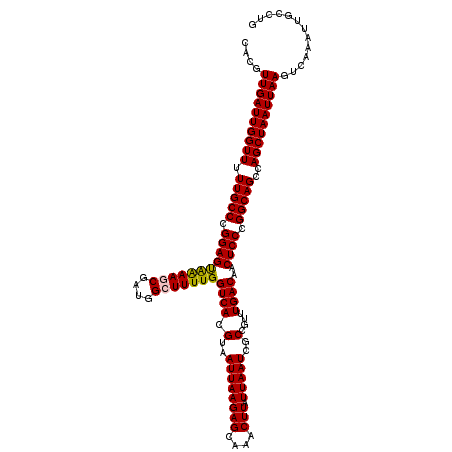
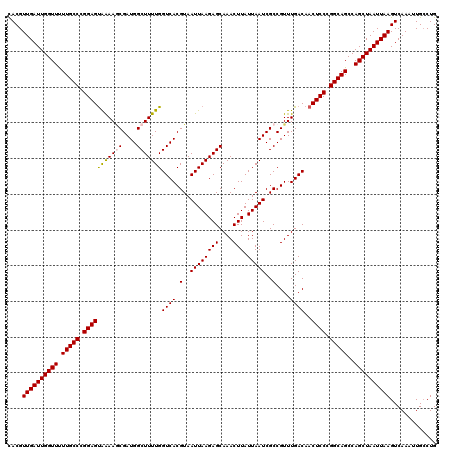
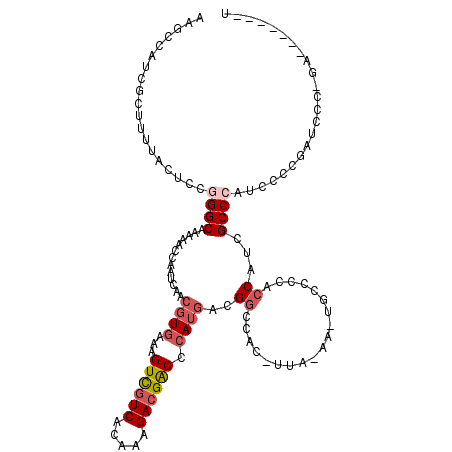
| Location | 1,324,271 – 1,324,391 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.99 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1324271 120 + 20766785 AAGCCAUCGCUUUUACUCCGGGCAAAAACCAAUCAACGUGAAAGUCGUCACAAAGACGACCCAUGACGGUCACCUUAAAAAUGCCCCACCAUCGCCCAUCCCCGAUCCCAGAUCCCCGAU .....((((..........((((.............((((...((((((.....)))))).))))..(((.................)))...))))......((((...))))..)))) ( -24.33) >DroSec_CAF1 42810 104 + 1 AAGCCAUCGCUUCUACUCCGGGCAAAAACCAAUCAACGUGAAAGUCGUCACAAAGACGACCCAUGACG-G---------------CCACCAUCGCCCAUCCCCGAUCCCCGAUCCUCGAU .....((((..........((((.............((((...((((((.....)))))).))))..(-(---------------...))...))))......((((...))))..)))) ( -23.20) >DroSim_CAF1 21891 98 + 1 AAGCCAUCGCUUCUACUCCGGGCAAAAACCAAUCAACGUGAAAGUCGUCACAAAGACGACCCAUGACGGG---------------CUACCAUCGCCCAUCGCCCAUCCCCGA-------U ((((....)))).......((((.............((((...((((((.....)))))).))))..(((---------------(.......))))...))))........-------. ( -29.50) >DroEre_CAF1 24160 105 + 1 AAGCCAUCGUUUUUGCUCCAGGCAAGAACCAAUCAACGUGAAAGUCCUCACAAAGACGACCCAA-CCGGCCACGUUAGAAGUGCCCCACCACUGCCCAUUCCGGAU-------------- ...((...((((((((.....))))))))...((((((((...(((.((.....)).)))((..-..)).)))))).))((((......)))).........))..-------------- ( -23.10) >DroYak_CAF1 10246 106 + 1 AAGCCAUCGAUUUCACUCCAGGCAAAAACCAAUCAACGUAAAAGUUGUCACAAAGACGGCCCAUGACGGCCACGUUAAAAAUGCCCCACCAUUGCCAAUUCCGGAU-------------- ...((...(....)......(((...........(((((.......(((.....)))((((......)))))))))...((((......)))))))......))..-------------- ( -19.90) >consensus AAGCCAUCGCUUUUACUCCGGGCAAAAACCAAUCAACGUGAAAGUCGUCACAAAGACGACCCAUGACGGCCAC_UUA_AA_UGCCCCACCAUCGCCCAUCCCCGAUCCC_GA_______U ...................((((.............((((...((((((.....)))))).))))..((...................))...))))....................... (-12.91 = -13.99 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:36 2006