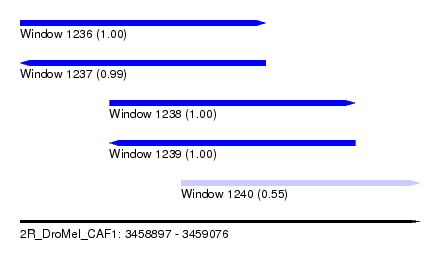
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,458,897 – 3,459,076 |
| Length | 179 |
| Max. P | 0.999453 |

| Location | 3,458,897 – 3,459,007 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.91 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
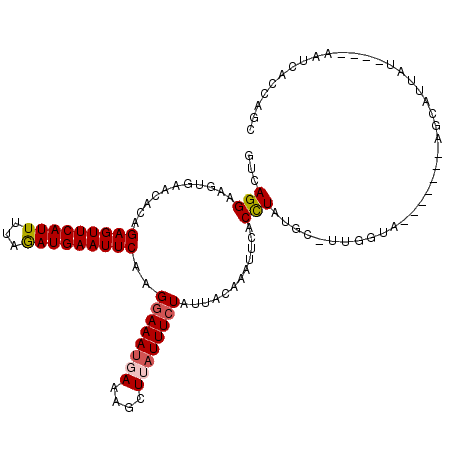
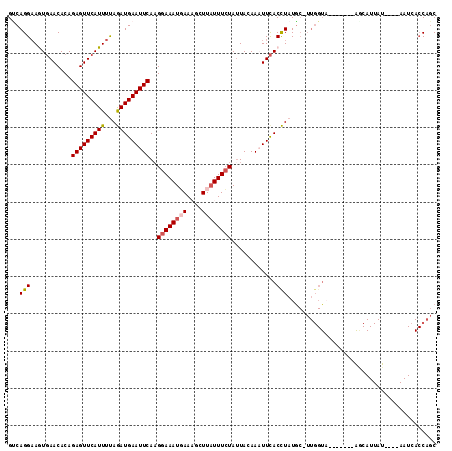
>2R_DroMel_CAF1 3458897 110 + 20766785 UUGGAAAAGAAGCAUUUUCUUUCUUUGGAUUCAUCAAUUAAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGCCAGGAAGUGAACACAGAGUUCAUUUCAGAUGAAUUC ..((((((......))))))......(((((((((......((((((((((((((((((.....(--------(.((....)).))...))))))))))))))))))..))))))))) ( -38.80) >DroSec_CAF1 9977 109 + 1 UUGGAA-AUAAGCAUUUUCUUUGUUUGAAUUAACCAAUUGAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUGAACACAGAGUUCAUUUUAGAUGAAUUC .(((..-.((((((.......))))))......)))(((.(((((((((((((((((((.(((..--------............))).))))))))))))))))))).)))...... ( -30.94) >DroSim_CAF1 12788 109 + 1 UUGGAA-AUAAGCAUUUUCUUUGUUUGGAUUAACCAAUUGAAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUUAACACAGAGUUCAUUUUAGAUGAAUUC .(((..-.((((((.......))))))......)))(((.(((((((((((((((((.((((..(--------(.((....)).)).))))))))))))))))))))).)))...... ( -29.80) >DroEre_CAF1 9271 107 + 1 UAAGAA-AUUAGCAUUCUCUUUGUUUGGAUAAACCAAUUGAAAAUGAGCUCUGUGUUUAAUUC---------GUAGAAGUGACAGG-AGUGAACACAGAGUUCAUUUUAGAUGAAUUC ......-......(((((((....((((.....))))...(((((((((((((((((((.(((---------............))-).)))))))))))))))))))))).)))).. ( -31.40) >DroYak_CAF1 9568 115 + 1 --AGAA-AUUAGCAUGUUCUUUGUUUGAAUGAACCAAUUGAGAAUGAGCUCUGUGUUGAAUUCGCAGAAUUCGCAGAAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAAAUGAAUUC --....-.....((((((((((....))).))))......(((((((((((((((((.(.(((((.......)).((....))..))).).)))))))))))))))))..)))..... ( -29.60) >consensus UUGGAA_AUAAGCAUUUUCUUUGUUUGGAUUAACCAAUUGAAAAUGAGCUCUGUGUUUAAUUCGC________CAGGAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAGAUGAAUUC ..........................(((((.....(((.(((((((((((((((((((.(((......................))).))))))))))))))))))).))).))))) (-24.27 = -24.91 + 0.64)


| Location | 3,458,897 – 3,459,007 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.93 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3458897 110 - 20766785 GAAUUCAUCUGAAAUGAACUCUGUGUUCACUUCCUGGCACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUUAAUUGAUGAAUCCAAAGAAAGAAAAUGCUUCUUUUCCAA (.(((((((.(((((((.(((((((((.(.((((..(.....)..--------).))).).))))))))).))))))).....))))))).)...(((((((.....))))))).... ( -33.00) >DroSec_CAF1 9977 109 - 1 GAAUUCAUCUAAAAUGAACUCUGUGUUCAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUCAAUUGGUUAAUUCAAACAAAGAAAAUGCUUAU-UUCCAA (((((.(((.(((((((.(((((((((...(((((.........)--------).)))...))))))))).))))))).....))).)))))...................-...... ( -24.00) >DroSim_CAF1 12788 109 - 1 GAAUUCAUCUAAAAUGAACUCUGUGUUAAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUUCAAUUGGUUAAUCCAAACAAAGAAAAUGCUUAU-UUCCAA ...(((....(((((((.((((((((((((((((.(......).)--------).))))).))))))))).)))))))...((((.....)))).....))).........-...... ( -25.50) >DroEre_CAF1 9271 107 - 1 GAAUUCAUCUAAAAUGAACUCUGUGUUCACU-CCUGUCACUUCUAC---------GAAUUAAACACAGAGCUCAUUUUCAAUUGGUUUAUCCAAACAAAGAGAAUGCUAAU-UUCUUA ..((((.((((((((((.(((((((((....-....((........---------))....))))))))).)))))))...((((.....))))....)))))))......-...... ( -22.80) >DroYak_CAF1 9568 115 - 1 GAAUUCAUUUAAAAUGAACUCUGUGUUCACUUCCUGACACUUCUGCGAAUUCUGCGAAUUCAACACAGAGCUCAUUCUCAAUUGGUUCAUUCAAACAAAGAACAUGCUAAU-UUCU-- ............(((((.(((((((((((.....))).........((((((...)))))).)))))))).)))))...(((((((...(((.......)))...))))))-)...-- ( -23.70) >consensus GAAUUCAUCUAAAAUGAACUCUGUGUUCACUUCCUGACACUCCUG________GCGAAUUAAACACAGAGCUCAUUUUCAAUUGGUUAAUCCAAACAAAGAAAAUGCUUAU_UUCCAA ...(((....(((((((.(((((((((...(((......................)))...))))))))).)))))))...((((.....)))).....)))................ (-17.81 = -17.93 + 0.12)

| Location | 3,458,937 – 3,459,047 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.49 |
| Mean single sequence MFE | -33.21 |
| Consensus MFE | -26.77 |
| Energy contribution | -28.13 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3458937 110 + 20766785 AAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGCCAGGAAGUGAACACAGAGUUCAUUUCAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUAAAAAUUCACCUAUG .((((((((((((((((((.....(--------(.((....)).))...)))))))))))))))))).((.((((((...((((((((....))))))))......)))))).))... ( -36.40) >DroSec_CAF1 10016 110 + 1 AAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUGAACACAGAGUUCAUUUUAGAUGAAUUCAUGGAAAUUAAAGCUUAUUUCUAUUACAAAUUCACCUAUG (((((((((((((((((((.(((..--------............))).)))))))))))))))))))((.((((((.((((((((........))))))))....)))))).))... ( -35.14) >DroSim_CAF1 12827 110 + 1 AAAAUGAGCUCUGUGUUUAAUUCGC--------CAGGAGUGUCAGGAAAUUAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAAUACAAAUUUACCUAUG (((((((((((((((((.((((..(--------(.((....)).)).)))))))))))))))))))))((.((((((...((((((((....))))))))......)))))).))... ( -30.00) >DroEre_CAF1 9310 108 + 1 AAAAUGAGCUCUGUGUUUAAUUC---------GUAGAAGUGACAGG-AGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAACGCUUAUUUCUAUUAUAAUUUCACCUAUG (((((((((((((((((((.(((---------............))-).)))))))))))))))))))((.((((.....((((((((....))))))))........)))).))... ( -33.02) >DroYak_CAF1 9605 118 + 1 AGAAUGAGCUCUGUGUUGAAUUCGCAGAAUUCGCAGAAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAAAUGAAUUCAAGGAAAUUAACGCUUUUUUGUAUUACAAUUUCACUUAUA (((((((((((((((((.(.(((((.......)).((....))..))).).)))))))))))))))))..........(((((((((((..(......)..)))..))))).)))... ( -31.50) >consensus AAAAUGAGCUCUGUGUUUAAUUCGC________CAGGAGUGUCAGGAAGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUACAAAUUCACCUAUG (((((((((((((((((((.(((......................))).)))))))))))))))))))((.((((((...((((((((....))))))))......)))))).))... (-26.77 = -28.13 + 1.36)

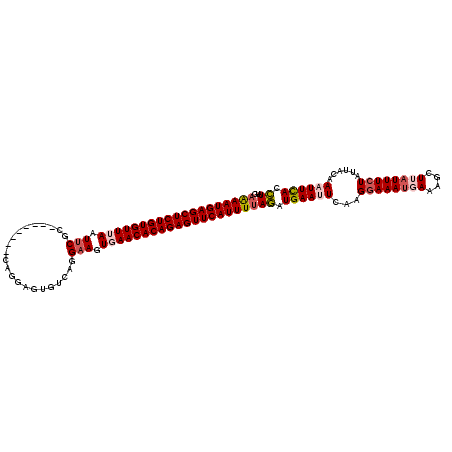
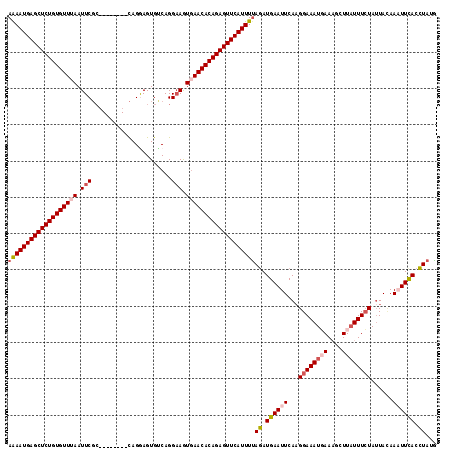
| Location | 3,458,937 – 3,459,047 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.49 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.79 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3458937 110 - 20766785 CAUAGGUGAAUUUUUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUGAAAUGAACUCUGUGUUCACUUCCUGGCACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUU ..(((((((((((......(((((........)))))...)))))))))))((((((.(((((((((.(.((((..(.....)..--------).))).).))))))))).)))))). ( -32.50) >DroSec_CAF1 10016 110 - 1 CAUAGGUGAAUUUGUAAUAGAAAUAAGCUUUAAUUUCCAUGAAUUCAUCUAAAAUGAACUCUGUGUUCAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUU ..((((((((((..(....(((((........))))).)..))))))))))((((((.(((((((((...(((((.........)--------).)))...))))))))).)))))). ( -31.80) >DroSim_CAF1 12827 110 - 1 CAUAGGUAAAUUUGUAUUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUAAUUUCCUGACACUCCUG--------GCGAAUUAAACACAGAGCUCAUUUU ..(((((.((((((.....(((((........)))))..)))))).)))))((((((.((((((((((((((((.(......).)--------).))))).))))))))).)))))). ( -26.10) >DroEre_CAF1 9310 108 - 1 CAUAGGUGAAAUUAUAAUAGAAAUAAGCGUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCACU-CCUGUCACUUCUAC---------GAAUUAAACACAGAGCUCAUUUU ..((((((((.(((.....(((((........)))))..))).))))))))((((((.(((((((((....-....((........---------))....))))))))).)))))). ( -24.80) >DroYak_CAF1 9605 118 - 1 UAUAAGUGAAAUUGUAAUACAAAAAAGCGUUAAUUUCCUUGAAUUCAUUUAAAAUGAACUCUGUGUUCACUUCCUGACACUUCUGCGAAUUCUGCGAAUUCAACACAGAGCUCAUUCU ..((((.(((((((................)))))))))))...........(((((.(((((((((((.....))).........((((((...)))))).)))))))).))))).. ( -24.49) >consensus CAUAGGUGAAUUUGUAAUAGAAAUAAGCUUUCAUUUCCUUGAAUUCAUCUAAAAUGAACUCUGUGUUCACUUCCUGACACUCCUG________GCGAAUUAAACACAGAGCUCAUUUU ..((((((((((((.....(((((........)))))..))))))))))))((((((.(((((((((...(((......................)))...))))))))).)))))). (-22.79 = -23.79 + 1.00)

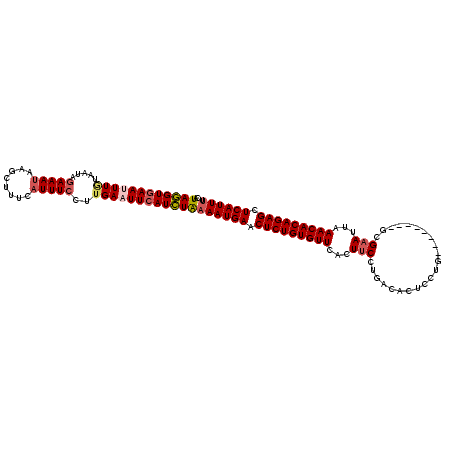
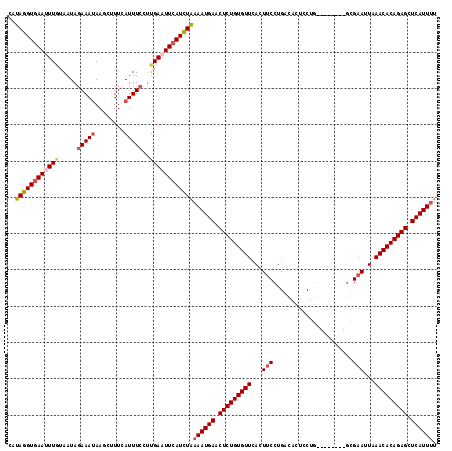
| Location | 3,458,969 – 3,459,076 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -12.52 |
| Energy contribution | -13.00 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3458969 107 + 20766785 GCCAGGAAGUGAACACAGAGUUCAUUUCAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUAAAAAUUCACCUAUGC-UUGGUA-------AGCAUUAUAUGUAAUAACCAGC ((((((..(((((....(((((((((...)))))))))..((((((((....))))))))........))))).....)-))))).-------...................... ( -24.50) >DroSec_CAF1 10048 104 + 1 GUCAGGAAAUGAACACAGAGUUCAUUUUAGAUGAAUUCAUGGAAAUUAAAGCUUAUUUCUAUUACAAAUUCACCUAUGCCUUGGUA-------AGCAUUAU----AAUCACCAGC ...(((((((((((.....))))))))(((.((((((.((((((((........))))))))....)))))).)))..)))((((.-------........----....)))).. ( -20.02) >DroSim_CAF1 12859 111 + 1 GUCAGGAAAUUAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAAUACAAAUUUACCUAUGCCUUGGUUUUACCUUGCCAUUAU----AAUCACCAGC ...((((((((......(((((((((...)))))))))..((((((((....))))))))......))))).)))......((((........))))....----.......... ( -20.60) >DroEre_CAF1 9341 81 + 1 GACAGG-AGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAACGCUUAUUUCUAUUAUAAUUUCACCUAUG---------------------------------CAAC ....((-.(((((....(((((((((...)))))))))..((((((((....))))))))........)))))))...---------------------------------.... ( -18.30) >DroYak_CAF1 9645 103 + 1 GUCAGGAAGUGAACACAGAGUUCAUUUUAAAUGAAUUCAAGGAAAUUAACGCUUUUUUGUAUUACAAUUUCACUUAUAC-UUGGUA-------AACAUUAU----AAUCACCAGC .(((.(((((((((.....)))))))))...)))....(((((((((((..(......)..)))..))))).)))....-(((((.-------........----....))))). ( -17.22) >consensus GUCAGGAAGUGAACACAGAGUUCAUUUUAGAUGAAUUCAAGGAAAUGAAAGCUUAUUUCUAUUACAAAUUCACCUAUGC_UUGGUA_______AGCAUUAU____AAUCACCAGC ...(((...........(((((((((...)))))))))..((((((((....))))))))............)))........................................ (-12.52 = -13.00 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:06 2006