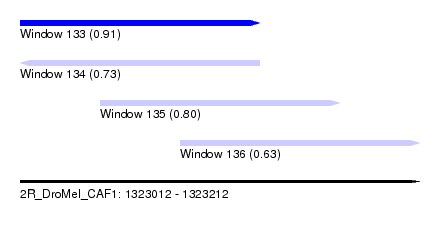
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,323,012 – 1,323,212 |
| Length | 200 |
| Max. P | 0.908858 |

| Location | 1,323,012 – 1,323,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -27.28 |
| Energy contribution | -30.04 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
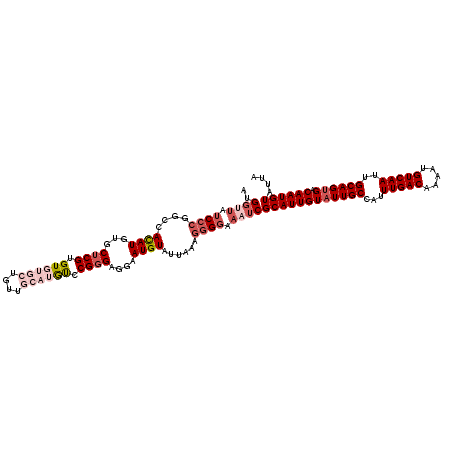
>2R_DroMel_CAF1 1323012 120 + 20766785 AUGGGUGUCCCGGCCACAUGUGCUCGUGUGUGCUGUUGCAUGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUA ...(((.(((((((((((((....)))))).)))..(((((.(((....))))))))......)))).)))((((((((((((...(((((.....)))))..))))).))))))).... ( -45.40) >DroSec_CAF1 41585 120 + 1 AUGGUUAUCCCGGCCACAUGUGCGCGUGUGUGCUGUUGCAUGUCCGGGAGGAAUGUAUUAAAUGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUA ..((((.(((((((((((((....)))))).)))..(((((.(((....))))))))......))))))))((((((((((((...(((((.....)))))..))))).))))))).... ( -45.80) >DroSim_CAF1 20608 120 + 1 AUGGUUAUCCCGGCCACAUGUGCUCGUGUGUGCUGUUGCAUGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUA ..((((.(((((((((((((....)))))).)))..(((((.(((....)))))))).....)))).))))((((((((((((...(((((.....)))))..))))).))))))).... ( -45.60) >DroEre_CAF1 22899 112 + 1 AUGCCAUACCCGGCCACAUGUGCUCGUGUA-------GCCCACGCGGG-GGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGCGACAAUGUAUUA ........(((....((((.(.(((((((.-------....)))))))-.).))))......)))......(((((((.((((...(((((.....)))))..))))..))))))).... ( -32.70) >DroYak_CAF1 8971 113 + 1 AUGGUUUUCACAGCCAUAUGUGCUCGUGUA-------GCACACACGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUA ..(((((((......((((((.(((((((.-------....)))))))....))))))......)))))))((((((((((((...(((((.....)))))..))))).))))))).... ( -35.10) >consensus AUGGUUAUCCCGGCCACAUGUGCUCGUGUGUGCUGUUGCAUGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUA ..((((.((((....((((...((((.((((((....)))))).))))....))))......)))).))))((((((((((((...(((((.....)))))..)))))).)))))).... (-27.28 = -30.04 + 2.76)

| Location | 1,323,012 – 1,323,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.94 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -20.33 |
| Energy contribution | -20.29 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1323012 120 - 20766785 UAAUACAUUGUCACUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCCUUUAAUACAUUCCUCCCGGACAUGCAACAGCACACACGAGCACAUGUGGCCGGGACACCCAU .....((((((...(((..(((((.....)))))...)))..)))))).......................((((((.(.(((....)))((((........)))))))))))....... ( -28.90) >DroSec_CAF1 41585 120 - 1 UAAUACAUUGUCACUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCAUUUAAUACAUUCCUCCCGGACAUGCAACAGCACACACGCGCACAUGUGGCCGGGAUAACCAU .....((((((...(((..(((((.....)))))...)))..)))))).......................(((((((((((.....((......))...)))))..))))))....... ( -29.40) >DroSim_CAF1 20608 120 - 1 UAAUACAUUGUCACUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCCUUUAAUACAUUCCUCCCGGACAUGCAACAGCACACACGAGCACAUGUGGCCGGGAUAACCAU .....((((((...(((..(((((.....)))))...)))..)))))).......................((((((.(.(((....)))((((........)))))))))))....... ( -29.00) >DroEre_CAF1 22899 112 - 1 UAAUACAUUGUCGCUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCCUUUAAUACAUUCC-CCCGCGUGGGC-------UACACGAGCACAUGUGGCCGGGUAUGGCAU .....((((((...(((..(((((.....)))))...)))..)))))).................(((.((-((((((((.((-------(.....))).)))))))..))).))).... ( -32.90) >DroYak_CAF1 8971 113 - 1 UAAUACAUUGUCACUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCCUUUAAUACAUUCCUCCCGUGUGUGC-------UACACGAGCACAUAUGGCUGUGAAAACCAU .....((((((...(((..(((((.....)))))...)))..))))))...................((((..((((((((((-------(.....)))))))))))..).)))...... ( -31.50) >consensus UAAUACAUUGUCACUGCAAUUGACAUUUUGUCAAAUGGCAAUACAAUGCGAUUUCCCCUUUAAUACAUUCCUCCCGGACAUGCAACAGCACACACGAGCACAUGUGGCCGGGAAAACCAU .....((((((...(((..(((((.....)))))...)))..)))))).......................(((((((((((..................)))))..))))))....... (-20.33 = -20.29 + -0.04)

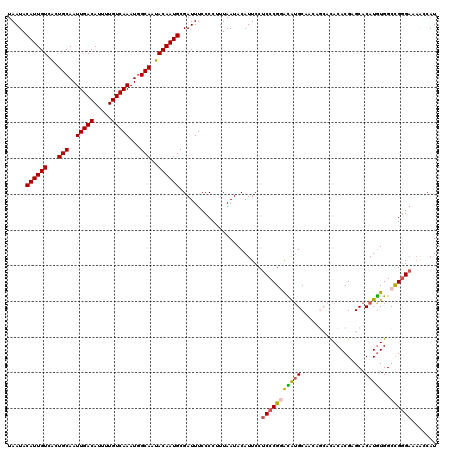
| Location | 1,323,052 – 1,323,172 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -27.40 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
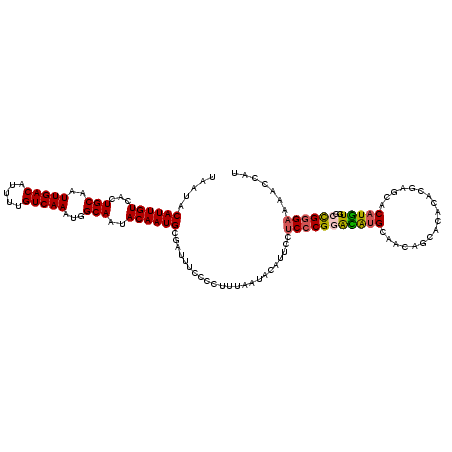
>2R_DroMel_CAF1 1323052 120 + 20766785 UGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGUACGGGCACAAUGGAGCGA (((((((((((((((((.....((.....))((((((((((((...(((((.....)))))..))))).))))))).........)))))...))))))....))))))........... ( -33.50) >DroSec_CAF1 41625 120 + 1 UGUCCGGGAGGAAUGUAUUAAAUGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGA (((((((((((((((((......((....))((((((((((((...(((((.....)))))..))))).))))))).........)))))...))))))....))))))........... ( -33.30) >DroSim_CAF1 20648 120 + 1 UGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGA (((((((((((((((((.....((.....))((((((((((((...(((((.....)))))..))))).))))))).........)))))...))))))....))))))........... ( -33.50) >DroEre_CAF1 22932 119 + 1 CACGCGGG-GGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGCGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGCGCACAAUGGAGCGA ..((((((-.(((((((.....((.....))(((((((.((((...(((((.....)))))..))))..))))))).........))))))).)))((((((....))....))))))). ( -31.10) >DroYak_CAF1 9004 120 + 1 CACACGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCAAGCACGGGCACAAUGGAGCGA .....((((((((((((.....((.....))((((((((((((...(((((.....)))))..))))).))))))).........))))))).)))))..((.(.(......).).)).. ( -27.80) >consensus UGUCCGGGAGGAAUGUAUUAAAGGGGAAAUCGCAUUGUAUUGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGA .((((((((((((((((......((....))((((((((((((...(((((.....)))))..)))))).)))))).........)))))...))))))....)))))............ (-27.40 = -28.12 + 0.72)

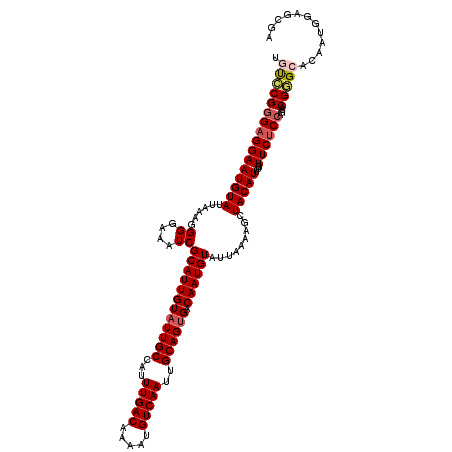
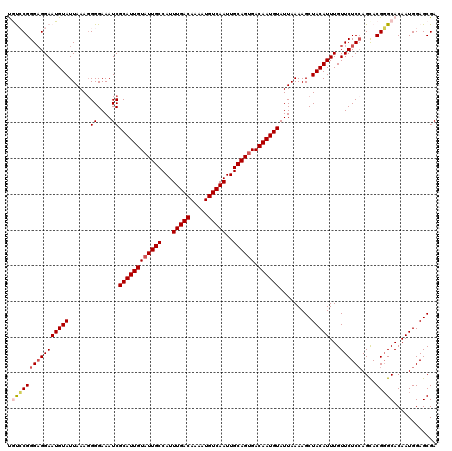
| Location | 1,323,092 – 1,323,212 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -26.38 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1323092 120 + 20766785 UGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGUACGGGCACAAUGGAGCGAUUUGCCAGCUGUCGAACAGAGGUACAAACAAGGACAUCCA (((...(((((.....)))))..))).................(.(((.(((((((..((((...((((.(((......))))))).))))..))))))).))))......((....)). ( -28.60) >DroSec_CAF1 41665 120 + 1 UGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGAUUUGCCAGCUGGCGAACAGAGGUACAAACAAGGAUAUCCA (((...(((((.....)))))..))).................(.(((.(((((((.(((((...((((.(((......))))))).))))).))))))).))))......((....)). ( -33.70) >DroSim_CAF1 20688 120 + 1 UGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGAUUUGCCAGCUGGCGAACAGAGGUACAAACAAGGAUAUCCA (((...(((((.....)))))..))).................(.(((.(((((((.(((((...((((.(((......))))))).))))).))))))).))))......((....)). ( -33.70) >DroEre_CAF1 22971 120 + 1 UGCCAUUUGACAAAAUGUCAAUUGCAGCGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGCGCACAAUGGAGCGAUUUGCCAGCUGCCGAGCAGAGGCAGCAACAAGGAUAUCCA (((...(((((.....)))))..)))..(((((((((........)))).))))).(((.(((..(((........)))...)))..((((((.......)))))).....)))...... ( -33.30) >DroYak_CAF1 9044 120 + 1 UGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCAAGCACGGGCACAAUGGAGCGAUUUGCCAGCUGCCGAACAGAGGUUCAAUCAAGGACAUCCA (((...(((((.....)))))..))).....((((((........))))))(((((...(((...((((.(((......))))))).)))...)))))((.((((......)))).)).. ( -26.50) >consensus UGCCAUUUGACAAAAUGUCAAUUGCAGUGACAAUGUAUUAAAAGCUACAUUUGUUCUCCAGCACGGGCACAAUGGAGCGAUUUGCCAGCUGCCGAACAGAGGUACAAACAAGGAUAUCCA ((((..(((((.....))))).((((.......))))............(((((((..((((...((((.(((......))))))).))))..))))))))))).......((....)). (-26.38 = -26.50 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:34 2006