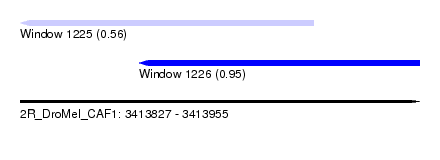
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,413,827 – 3,413,955 |
| Length | 128 |
| Max. P | 0.947144 |

| Location | 3,413,827 – 3,413,921 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -24.27 |
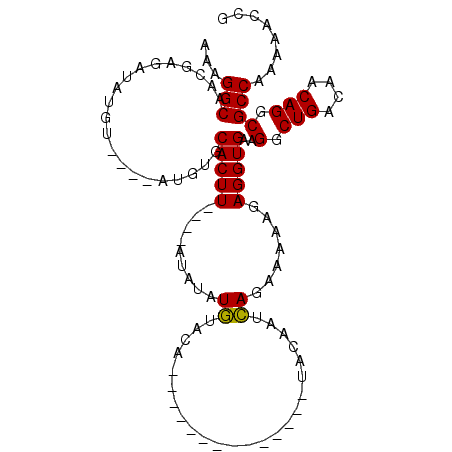
| Consensus MFE | -16.55 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3413827 94 - 20766785 AUGUGUACA--------------UACAAUCA-GAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCGCAUUGCUCGAUGCAGCAGCAGCCA-CACUGCAGCGGCAA .((((....--------------))))....-..............(.(((....))).)(((.......(((((....))))).((.((((...-..)))).))))).. ( -23.20) >DroSec_CAF1 44005 89 - 1 AUAUGUACA--------------UACAAUCA-GAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCGCAUUGCUCGAUGCAGCAGC------CACUGCAGCGGCAA .........--------------........-........((((....(((....))).)))).....((((..(((.....)))((((.------..)))).))))... ( -20.10) >DroSim_CAF1 44209 89 - 1 AUAUGUACA--------------UACAAUCA-GAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCGCAUUGCUCGAUGCAGCAGC------CACUGCAGCGGCAA .........--------------........-........((((....(((....))).)))).....((((..(((.....)))((((.------..)))).))))... ( -20.10) >DroEre_CAF1 44823 106 - 1 AUAUCUACAUUCAUGCGCGAGCGUACGAAGA-GAAAAAGAGGUGAAGGCUGACGACAGGCGCCAAAAACCGCAUUGCUCGAUGCUGCAGC---CAUAACUGCAGCGGCAA ...(((...(((((((....))))..))).)-)).............(((..(((((((((........))).))).)))..(((((((.---.....)))))))))).. ( -30.90) >DroYak_CAF1 47319 103 - 1 AUAUCUACACUCAUGCGCAAGCGUACGAAGA-GAAAAAGAGGUGAAGGCUGACGACAGGCGCCAAAAACCGCAUUGUUCGAUGCAGCAGC------AACUGCAGCGGCAA .....(((.(((((((....)))).(.....-).....))))))..(.(((....))).)(((.......(((((....))))).((((.------..))))...))).. ( -28.50) >DroPer_CAF1 44966 83 - 1 ----CCACAU----------GCAUUCGGAGAGGAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCGCAUUGCU-GCUGCUGCA------------GCAGCAACCA ----.....(----------((.(((......))).....((((....(((....))).)))).......)))(((((-((((...))------------)))))))... ( -22.80) >consensus AUAUCUACA______________UACAAACA_GAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCGCAUUGCUCGAUGCAGCAGC______CACUGCAGCGGCAA ........................................((((....(((....))).)))).....((((...((.....)).((((.........)))).))))... (-16.55 = -16.95 + 0.40)

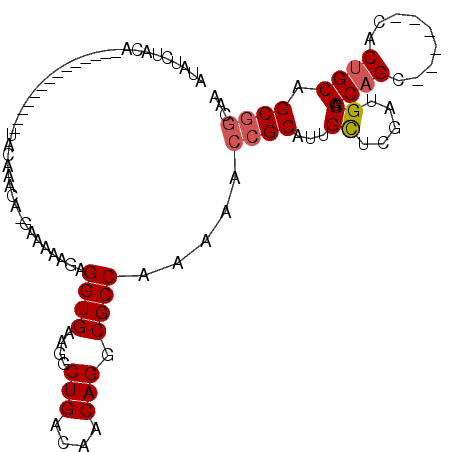
| Location | 3,413,865 – 3,413,955 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -24.74 |
| Consensus MFE | -11.09 |
| Energy contribution | -10.61 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3413865 90 - 20766785 AAAGGCAACGAGAUAUGUAUGUAUGUGCACUU----AUAUGUGUACA--------------UACAAUCAGAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCG ...(((.............(((((((((((..----....)))))))--------------))))..................(.(((....))).))))........ ( -25.70) >DroSec_CAF1 44038 86 - 1 AAAGGCAACGAGAUAUGU----AUGUGCACUU----GUAUAUGUACA--------------UACAAUCAGAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCG ...(((.....(((.(((----(((((((...----.....))))))--------------)))))))...............(.(((....))).))))........ ( -23.30) >DroSim_CAF1 44242 88 - 1 AAAGGCAACGAGAUAUGU--GCAUGUGCACUU----GUAUAUGUACA--------------UACAAUCAGAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCG ...(((...((..(((((--((((((((....----)))))))))))--------------))...))...............(.(((....))).))))........ ( -27.00) >DroEre_CAF1 44859 96 - 1 AAAGGCAACGAGAUAUG--------UGCACUU----AUAUAUCUACAUUCAUGCGCGAGCGUACGAAGAGAAAAAGAGGUGAAGGCUGACGACAGGCGCCAAAAACCG ...(((..(((((((((--------((....)----))))))))......((((....)))).))..................(.(((....))).))))........ ( -23.10) >DroYak_CAF1 47352 100 - 1 AAAGGCAACGAGAUAUG--------UGCACUUACAUAUAUAUCUACACUCAUGCGCAAGCGUACGAAGAGAAAAAGAGGUGAAGGCUGACGACAGGCGCCAAAAACCG ...(((...((.(((((--------((......))))))).))(((.(((((((....)))).(.....).....))))))..(.(((....))).))))........ ( -24.60) >consensus AAAGGCAACGAGAUAUGU____AUGUGCACUU____AUAUAUGUACA______________UACAAUCAGAAAAAGAGGUGAAGGCUGACAACAGGCGCCAAAAACCG ...(((.....................(((((.........((........................)).......)))))..(.(((....))).))))........ (-11.09 = -10.61 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:52 2006