
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,412,266 – 3,412,412 |
| Length | 146 |
| Max. P | 0.988643 |

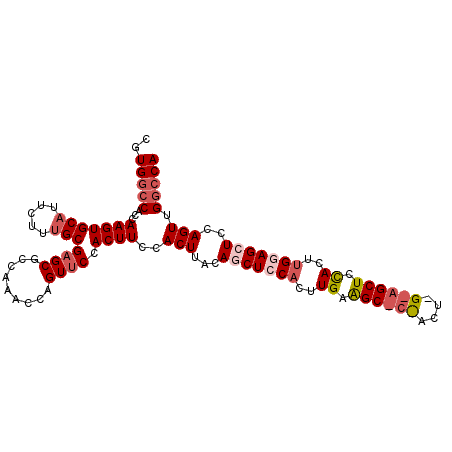
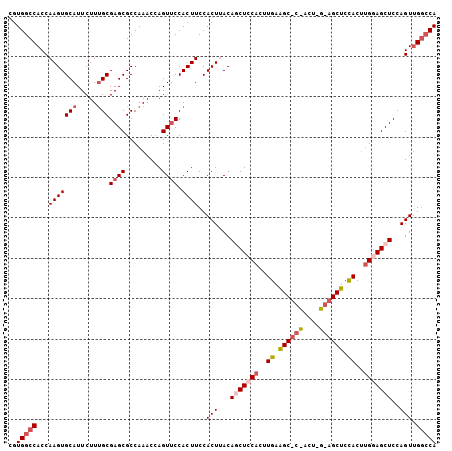
| Location | 3,412,266 – 3,412,372 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -19.60 |
| Energy contribution | -22.08 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
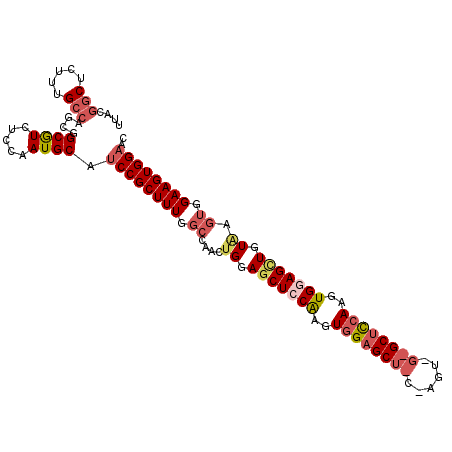
>2R_DroMel_CAF1 3412266 106 + 20766785 CGUGGCCACCAAGUGCAUUCUUUGCGAGCGCCAAACAAGUUCCACUUCCACUUACAACUCCACUUGAAGCACCACUCGCAGCUCCACUUGGAGCUCCAGUUGGCCA ..((((((.((((((((.....)))((((.........))))...................))))).......(((.(.((((((....))))))).))))))))) ( -30.40) >DroSec_CAF1 42452 82 + 1 CGUGGCCACCAAGUGCAUUCUUUGCGAGCGCCAAACCAGUUCCACUUCCACUUACAGCUCCACU------------------------UGGAGCUCCAGUUGGCCA ..((((((.((((((((.....)))((((.........)))).......))))..((((((...------------------------.))))))...).)))))) ( -26.20) >DroSim_CAF1 41693 106 + 1 CGUGGCCACCAAGUGCAUUCUUCGCGAGCGCCAAACCAGUUCCACUUCCACUUACAGCUCCACUUGGAGCUCCACUUGGAGCUCCACUUGGAGCUCCAGUUGGCCA ..((((((.(((((((.......))((((.........)))).......))))..(((((((..(((((((((....)))))))))..)))))))...).)))))) ( -45.20) >DroEre_CAF1 42973 93 + 1 CGUGGCCACCAAGUGCAUUCUUUGCGAGCGCCAAACCAGUUCCACUUCCACUCACACCUCCACUUGAGG------------CUCCACUCGCAGCUCCAGUGG-GCA .((((.....(((((((.....)))((((.........)))).)))))))).......((((((.(..(------------((.(....).)))..))))))-).. ( -22.30) >DroYak_CAF1 45619 106 + 1 UGUGGCCACCAAGUGCAUUCUUUGCGAGCGCCAAACCAGUGCCACUUCCACUAACAGCUCCACUUGGAGCUCUACUUGGAGCUCUACUUGCAGCUCCAGUUGGCCA ..(((((..((((((((.....)))((((.((((...((((.......))))...((((((....))))))....)))).)))).))))).(((....)))))))) ( -37.80) >consensus CGUGGCCACCAAGUGCAUUCUUUGCGAGCGCCAAACCAGUUCCACUUCCACUUACAGCUCCACUUGAAGC_C_ACU_G_AGCUCCACUUGGAGCUCCAGUUGGCCA ..(((((...(((((((.....)))((((.........)))).))))..(((...(((((((..((.((((((....)))))).))..)))))))..))).))))) (-19.60 = -22.08 + 2.48)

| Location | 3,412,266 – 3,412,372 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -30.05 |
| Energy contribution | -32.16 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3412266 106 - 20766785 UGGCCAACUGGAGCUCCAAGUGGAGCUGCGAGUGGUGCUUCAAGUGGAGUUGUAAGUGGAAGUGGAACUUGUUUGGCGCUCGCAAAGAAUGCACUUGGUGGCCACG (((((((((.(((((((....)))))).).)))......(((((((..(((.(..((((..((.(((....))).))..))))..).))).))))))))))))).. ( -39.80) >DroSec_CAF1 42452 82 - 1 UGGCCAACUGGAGCUCCA------------------------AGUGGAGCUGUAAGUGGAAGUGGAACUGGUUUGGCGCUCGCAAAGAAUGCACUUGGUGGCCACG ...(((..((.((((((.------------------------...)))))).))..)))..((((.((..(....(((.((.....)).)))..)..))..)))). ( -29.70) >DroSim_CAF1 41693 106 - 1 UGGCCAACUGGAGCUCCAAGUGGAGCUCCAAGUGGAGCUCCAAGUGGAGCUGUAAGUGGAAGUGGAACUGGUUUGGCGCUCGCGAAGAAUGCACUUGGUGGCCACG ...(((..((.(((((((..(((((((((....)))))))))..))))))).))..)))..((((.((..(....(((.((.....)).)))..)..))..)))). ( -52.60) >DroEre_CAF1 42973 93 - 1 UGC-CCACUGGAGCUGCGAGUGGAG------------CCUCAAGUGGAGGUGUGAGUGGAAGUGGAACUGGUUUGGCGCUCGCAAAGAAUGCACUUGGUGGCCACG ...-(((((.(.....).))))).(------------(((((((((.(..(((((((....((.(((....))).))))))))).....).))))))).))).... ( -32.70) >DroYak_CAF1 45619 106 - 1 UGGCCAACUGGAGCUGCAAGUAGAGCUCCAAGUAGAGCUCCAAGUGGAGCUGUUAGUGGAAGUGGCACUGGUUUGGCGCUCGCAAAGAAUGCACUUGGUGGCCACA ((((((.((((((((........))))))((((.((((.((((..((.((..((......))..)).))...)))).))))(((.....))))))))))))))).. ( -40.70) >consensus UGGCCAACUGGAGCUCCAAGUGGAGCU_C_AGU_G_GCUCCAAGUGGAGCUGUAAGUGGAAGUGGAACUGGUUUGGCGCUCGCAAAGAAUGCACUUGGUGGCCACG (((((.((..((((((((..((((((((......))))))))..)))))))....((((..((.(((....))).))..))))...........)..))))))).. (-30.05 = -32.16 + 2.11)

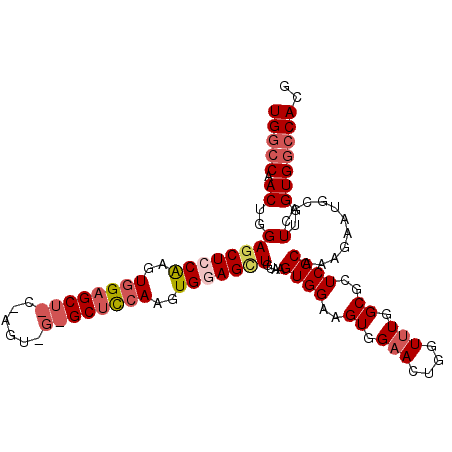
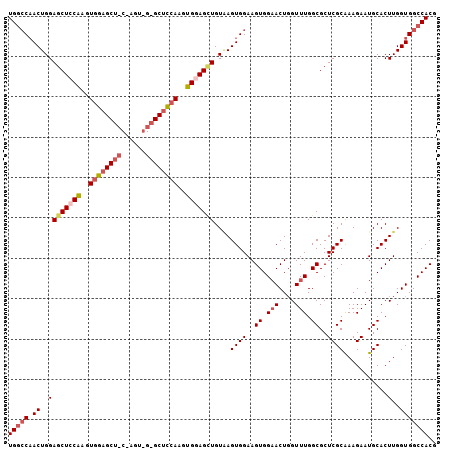
| Location | 3,412,304 – 3,412,412 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -30.61 |
| Energy contribution | -33.04 |
| Covariance contribution | 2.43 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3412304 108 - 20766785 UUACGGCUCUUUGCCAGCGGCGUCUCCAAUGCAUCCGCUUUGGCCAACUGGAGCUCCAAGUGGAGCUGCGAGUGGUGCUUCAAGUGGAGUUGUAAGUGGAAGUGGAAC ....(((.....)))....((((.....)))).(((((((...(((..((.(((((((..((((((.((.....))))))))..))))))).))..)))))))))).. ( -41.70) >DroSec_CAF1 42490 84 - 1 UUACGGCUCUUUGCCAGCGGCGUCUCCAAUGCAUCCGCUUUGGCCAACUGGAGCUCCA------------------------AGUGGAGCUGUAAGUGGAAGUGGAAC ....(((.....)))....((((.....)))).(((((((...(((..((.((((((.------------------------...)))))).))..)))))))))).. ( -32.00) >DroSim_CAF1 41731 107 - 1 NNNNNNCUC-UUGCCAGCGGCGUCUCCAAUGCAUCCGCUUUGGCCAACUGGAGCUCCAAGUGGAGCUCCAAGUGGAGCUCCAAGUGGAGCUGUAAGUGGAAGUGGAAC .........-..(((...)))............(((((((...(((..((.(((((((..(((((((((....)))))))))..))))))).))..)))))))))).. ( -50.00) >DroEre_CAF1 43011 95 - 1 UUACGGCUCUUUGCCAGCGGCGUCUCCAAUGCAUCCGCUUUGC-CCACUGGAGCUGCGAGUGGAG------------CCUCAAGUGGAGGUGUGAGUGGAAGUGGAAC ....(((.....)))....((((.....)))).((((((((..-(((((.(.....).))))).(------------((((.....))))).......)))))))).. ( -32.50) >DroYak_CAF1 45657 107 - 1 -UACGGCUCUUUGCCAGCGGCAUCUCCAAUGCAUCCGCUUUGGCCAACUGGAGCUGCAAGUAGAGCUCCAAGUAGAGCUCCAAGUGGAGCUGUUAGUGGAAGUGGCAC -...(((.....))).((.((((.....))))...((((((.((.((((((((((........))))))).....((((((....))))))))).)).)))))))).. ( -42.70) >consensus UUACGGCUCUUUGCCAGCGGCGUCUCCAAUGCAUCCGCUUUGGCCAACUGGAGCUCCAAGUGGAGCU_C_AGU_G_GCUCCAAGUGGAGCUGUAAGUGGAAGUGGAAC ....(((.....)))....((((.....)))).((((((((.((....((.(((((((..((((((((......))))))))..))))))).)).)).)))))))).. (-30.61 = -33.04 + 2.43)

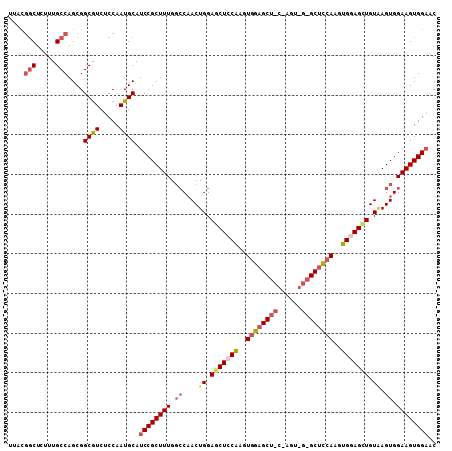
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:50 2006