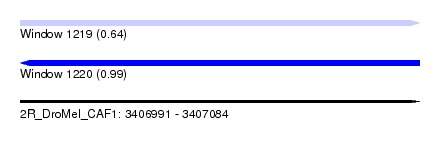
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,406,991 – 3,407,084 |
| Length | 93 |
| Max. P | 0.994620 |

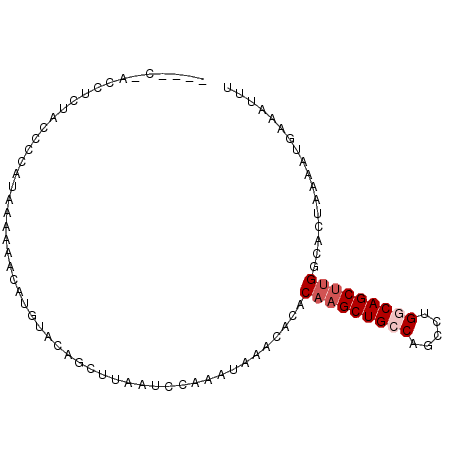
| Location | 3,406,991 – 3,407,084 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
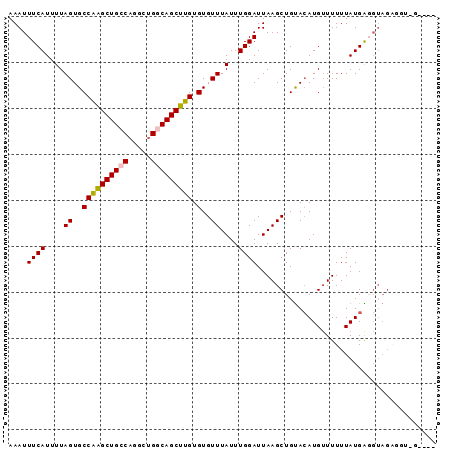
>2R_DroMel_CAF1 3406991 93 + 20766785 AAAUUUCAUUUUAGUGCCAAGCUGCCAGGCAGUCAGCUUGUGUGUUUAUUUGGAUUAAGCUGUACAUGUUUUUUAUGGUGUAGUUGUAGGGGG ....((((....((..((((((((.(.....).))))))).)..))....))))...(((((((((((.....))).))))))))........ ( -19.20) >DroSec_CAF1 37237 89 + 1 AAAUUUCAUUUUAGUGCCAAGCUGCCAGGCUGGCAGCUUGUGUGUUUAUUUGGAUUAAGCUGUACAUGUUUUUUAUGAGGUAGAGGUUG---- ....((((....((..((((((((((.....))))))))).)..))....)))).(((.((.(((..((.....))...))).)).)))---- ( -25.40) >DroSim_CAF1 36549 89 + 1 AAAUUUCAUUUUAGUGCCGAGCUGCCAGGCUGGCAGCUUGUGUGUUUAUUUGGAUUAAGCUGUACAUGUUUUUUAUGAGGUAGAGGGGG---- ....((((....((..((((((((((.....))))))))).)..))....))))....(((...((((.....)))).)))........---- ( -24.30) >DroYak_CAF1 40767 70 + 1 AAAUUUCAUUUUAGUGCCAGGCUGG--------CAGCUUGUGUGUUUAUUUGGAUUAAGCUGCAAAUGAUUUUUAUGG--------------- (((..((((((((((.....))))(--------(((((((...............))))))))))))))..)))....--------------- ( -15.76) >consensus AAAUUUCAUUUUAGUGCCAAGCUGCCAGGCUGGCAGCUUGUGUGUUUAUUUGGAUUAAGCUGUACAUGUUUUUUAUGAGGUAGAGGU_G____ ....((((....((..((((((((((.....))))))))).)..))....))))....................................... (-17.89 = -18.07 + 0.19)

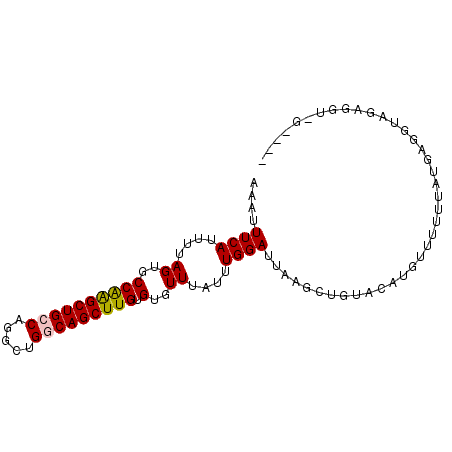
| Location | 3,406,991 – 3,407,084 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -13.38 |
| Consensus MFE | -11.09 |
| Energy contribution | -12.15 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3406991 93 - 20766785 CCCCCUACAACUACACCAUAAAAAACAUGUACAGCUUAAUCCAAAUAAACACACAAGCUGACUGCCUGGCAGCUUGGCACUAAAAUGAAAUUU .....................................................(((((((.((....)))))))))................. ( -11.30) >DroSec_CAF1 37237 89 - 1 ----CAACCUCUACCUCAUAAAAAACAUGUACAGCUUAAUCCAAAUAAACACACAAGCUGCCAGCCUGGCAGCUUGGCACUAAAAUGAAAUUU ----.................................................(((((((((.....)))))))))................. ( -17.40) >DroSim_CAF1 36549 89 - 1 ----CCCCCUCUACCUCAUAAAAAACAUGUACAGCUUAAUCCAAAUAAACACACAAGCUGCCAGCCUGGCAGCUCGGCACUAAAAUGAAAUUU ----.................................................(.(((((((.....))))))).)................. ( -13.30) >DroYak_CAF1 40767 70 - 1 ---------------CCAUAAAAAUCAUUUGCAGCUUAAUCCAAAUAAACACACAAGCUG--------CCAGCCUGGCACUAAAAUGAAAUUU ---------------.........(((((((((((((.................))))))--------)..((...))....))))))..... ( -11.53) >consensus ____C_ACCUCUACCCCAUAAAAAACAUGUACAGCUUAAUCCAAAUAAACACACAAGCUGCCAGCCUGGCAGCUUGGCACUAAAAUGAAAUUU .....................................................(((((((((.....)))))))))................. (-11.09 = -12.15 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:47 2006