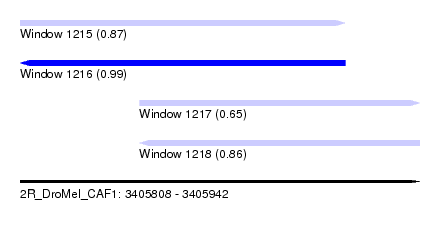
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,405,808 – 3,405,942 |
| Length | 134 |
| Max. P | 0.991442 |

| Location | 3,405,808 – 3,405,917 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -20.78 |
| Energy contribution | -24.00 |
| Covariance contribution | 3.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3405808 109 + 20766785 GCGGCCAGGCCCCAAAUUGAUUGGGUGACCGACCGACUGGUUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUCACGGCCAAUUAAUUGGCCAUUUCUCGUUGGCCCAAAUA- (.(((((((..(((..(((.((((....)))).))).)))..))((((..(((((.-....)))))..))))..((((((....)))))).........)))))).....- ( -41.90) >DroSec_CAF1 36042 109 + 1 GCGGCCAGGCCCCAAAUUGAUUGGGUGACCGACCGACUGGUUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUUGCCCAAAUA- (((((.((((((.(((((..((((........))))..))))).)))))).((((.-...)))).)))))....((((((....))))))....................- ( -40.40) >DroSim_CAF1 35345 109 + 1 GCGGCCAGGCCCCAAAUUGAUUGGGUGACCGACCGACUGGUUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA- (.(((((((((((.......((((....))))((((......)))).....)))))-.................((((((....))))))........))))))).....- ( -42.60) >DroEre_CAF1 36512 102 + 1 GCGGCCAGCCCCCAAAUUGAUUGGGUUA--------CUGGUUUCGGGCCUCUGGGUUUUUCCUCUGUCGCUUACUGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA- (..(((((..(((((.....)))))...--------)))))..)(((((...(((....))).............(((((....)))))...........))))).....- ( -32.20) >DroYak_CAF1 39613 102 + 1 GCGGCCAGGCCCCAAAUUGAUUGGGUUA--------CUGGUUGCGGGCCUCUGGGUUUUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA- ((((((((..(((((.....)))))...--------))))))))(((((...(((....)))............((((((....))))))..........))))).....- ( -45.40) >DroPer_CAF1 37720 90 + 1 GCAGCC------------GACUGGUUUG--------CUCUUGUCCAAUUUUCGGGU-UUCCCGCUUUCCAUUGUCUGCAUUUAAUAUGUAAUUCUCCGCCUGCCCAAAAAG ((((.(------------((((((...(--------(......((........)).-.....))...)))..)))(((((.....))))).......).))))........ ( -15.80) >consensus GCGGCCAGGCCCCAAAUUGAUUGGGUGA________CUGGUUUCGGGCCUCGGGGU_UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA_ (..(((((..(((((.....)))))...........)))))..)(((((...(((....)))............((((((....))))))..........)))))...... (-20.78 = -24.00 + 3.22)

| Location | 3,405,808 – 3,405,917 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -40.54 |
| Consensus MFE | -23.37 |
| Energy contribution | -26.07 |
| Covariance contribution | 2.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3405808 109 - 20766785 -UAUUUGGGCCAACGAGAAAUGGCCAAUUAAUUGGCCGUGAGCGACAGAGGGAA-ACCCCGAGGCCCGAAACCAGUCGGUCGGUCACCCAAUCAAUUUGGGGCCUGGCCGC -..((((((((..((....((((((((....))))))))...))...(.((...-.)).)..))))))))......(((((((.(.(((((.....)))))).))))))). ( -50.70) >DroSec_CAF1 36042 109 - 1 -UAUUUGGGCAAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA-ACCCCGAGGCCCGAAACCAGUCGGUCGGUCACCCAAUCAAUUUGGGGCCUGGCCGC -..(((((((..((.....((((((((....))))))))..))....(.((...-.)).)...)))))))......(((((((.(.(((((.....)))))).))))))). ( -46.40) >DroSim_CAF1 35345 109 - 1 -UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA-ACCCCGAGGCCCGAAACCAGUCGGUCGGUCACCCAAUCAAUUUGGGGCCUGGCCGC -..((((((((.((.....((((((((....))))))))..))....(.((...-.)).)..))))))))......(((((((.(.(((((.....)))))).))))))). ( -49.90) >DroEre_CAF1 36512 102 - 1 -UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCAGUAAGCGACAGAGGAAAAACCCAGAGGCCCGAAACCAG--------UAACCCAAUCAAUUUGGGGGCUGGCCGC -..((((((((.((........(((((....))))).....))......((......))...)))))))).((((--------(..(((((.....))))).))))).... ( -37.22) >DroYak_CAF1 39613 102 - 1 -UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAAAACCCAGAGGCCCGCAACCAG--------UAACCCAAUCAAUUUGGGGCCUGGCCGC -.....(((((.((.....((((((((....))))))))..))......(((....)))...)))))((..((((--------...(((((.....)))))..))))..)) ( -42.40) >DroPer_CAF1 37720 90 - 1 CUUUUUGGGCAGGCGGAGAAUUACAUAUUAAAUGCAGACAAUGGAAAGCGGGAA-ACCCGAAAAUUGGACAAGAG--------CAAACCAGUC------------GGCUGC ........(((((((...(((.....)))...))).(((..(((...((((...-.))(((...))).......)--------)...))))))------------..)))) ( -16.60) >consensus _UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA_ACCCCGAGGCCCGAAACCAG________UAACCCAAUCAAUUUGGGGCCUGGCCGC ...((((((((..((....((((((((....))))))))...)).....(((....)))...))))))))................(((((.....))))).......... (-23.37 = -26.07 + 2.70)

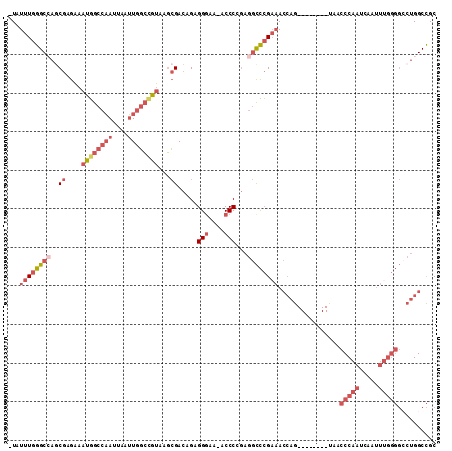
| Location | 3,405,848 – 3,405,942 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.57 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3405848 94 + 20766785 UUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUCACGGCCAAUUAAUUGGCCAUUUCUCGUUGGCCCAAAUA--CACGGACAUCCGGCCCCUUUGAC--------CG ....((((..(((((.-....)))))..))))..((((........(((((........)))))......--...((....))))))........--------.. ( -30.30) >DroSec_CAF1 36082 94 + 1 UUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUUGCCCAAAUA--CACGGACAUCCGGCCCCUUUGAC--------CU ....(((((...((((-........((.....))((((((....))))))...........)))).....--...((....))))))).......--------.. ( -27.10) >DroSim_CAF1 35385 94 + 1 UUUCGGGCCUCGGGGU-UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA--CACGGACAUCCGGCCCCUUUGAC--------CU ....(((((..((((.-...))))..........((((((....))))))..........))))).....--..(((....)))...........--------.. ( -29.40) >DroEre_CAF1 36544 95 + 1 UUUCGGGCCUCUGGGUUUUUCCUCUGUCGCUUACUGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA--CACGGACAUCCGGCCCCUUUGAC--------CG ....(((((...(((....)))..((((((.....)).........(((((........)))))......--....))))...))))).......--------.. ( -25.30) >DroYak_CAF1 39645 95 + 1 UUGCGGGCCUCUGGGUUUUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA--CACGGACAUCCGGCCCCUUUGAC--------UG ..(((((((...(((....)))............((((((....))))))..........))))).....--..(((....))))).........--------.. ( -30.60) >DroPer_CAF1 37740 104 + 1 UGUCCAAUUUUCGGGU-UUCCCGCUUUCCAUUGUCUGCAUUUAAUAUGUAAUUCUCCGCCUGCCCAAAAAGUCACGGAUGUCCGGCCCCACCGCCACCGCCACCG .((((..((((.((((-.....((........)).(((((.....)))))...........)))).)))).....))))....(((......))).......... ( -15.40) >consensus UUUCGGGCCUCGGGGU_UUCCCUCUGUCGCUUACGGCCAAUUAAUUGGCCAUUUCUCGCUGGCCCAAAUA__CACGGACAUCCGGCCCCUUUGAC________CG ....(((((...(((....)))..((((((.....)).........(((((........)))))............))))...)))))................. (-20.87 = -20.57 + -0.30)

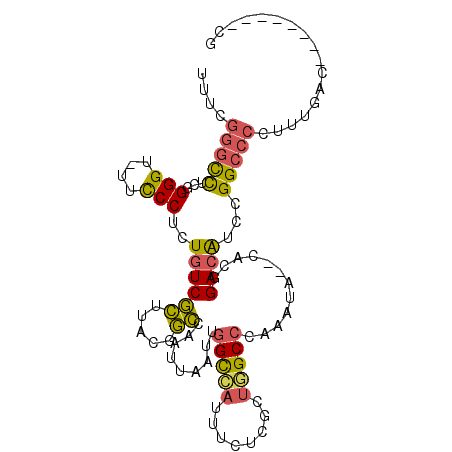
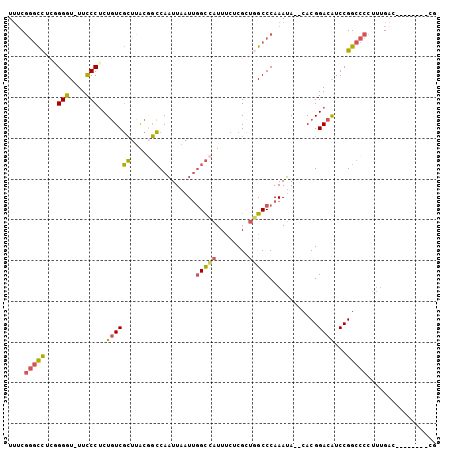
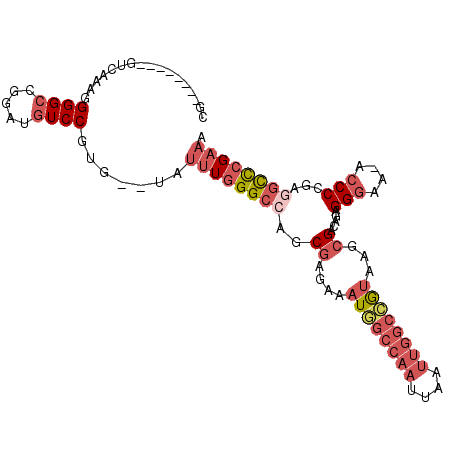
| Location | 3,405,848 – 3,405,942 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -22.95 |
| Energy contribution | -24.98 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3405848 94 - 20766785 CG--------GUCAAAGGGGCCGGAUGUCCGUG--UAUUUGGGCCAACGAGAAAUGGCCAAUUAAUUGGCCGUGAGCGACAGAGGGAA-ACCCCGAGGCCCGAAA .(--------(((.....))))((....))...--..((((((((..((....((((((((....))))))))...))...(.((...-.)).)..)))))))). ( -39.70) >DroSec_CAF1 36082 94 - 1 AG--------GUCAAAGGGGCCGGAUGUCCGUG--UAUUUGGGCAAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA-ACCCCGAGGCCCGAAA ..--------.......(((((...((((((..--....)))))).((.....((((((((....))))))))..))....(.((...-.)).)..))))).... ( -36.20) >DroSim_CAF1 35385 94 - 1 AG--------GUCAAAGGGGCCGGAUGUCCGUG--UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA-ACCCCGAGGCCCGAAA .(--------(((.....))))((....))...--..((((((((.((.....((((((((....))))))))..))....(.((...-.)).)..)))))))). ( -38.80) >DroEre_CAF1 36544 95 - 1 CG--------GUCAAAGGGGCCGGAUGUCCGUG--UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCAGUAAGCGACAGAGGAAAAACCCAGAGGCCCGAAA .(--------(((.....))))((....))...--..((((((((.((........(((((....))))).....))......((......))...)))))))). ( -30.62) >DroYak_CAF1 39645 95 - 1 CA--------GUCAAAGGGGCCGGAUGUCCGUG--UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAAAACCCAGAGGCCCGCAA ..--------.........(((((....))).)--)....(((((.((.....((((((((....))))))))..))......(((....)))...))))).... ( -33.60) >DroPer_CAF1 37740 104 - 1 CGGUGGCGGUGGCGGUGGGGCCGGACAUCCGUGACUUUUUGGGCAGGCGGAGAAUUACAUAUUAAAUGCAGACAAUGGAAAGCGGGAA-ACCCGAAAAUUGGACA .(((.((((((.(((.....)))..)).)))).)))..........(((...(((.....)))...)))...((((.....(.((...-.)))....)))).... ( -24.40) >consensus CG________GUCAAAGGGGCCGGAUGUCCGUG__UAUUUGGGCCAGCGAGAAAUGGCCAAUUAAUUGGCCGUAAGCGACAGAGGGAA_ACCCCGAGGCCCGAAA .................((((.....)))).......((((((((..((....((((((((....))))))))...)).....(((....)))...)))))))). (-22.95 = -24.98 + 2.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:45 2006