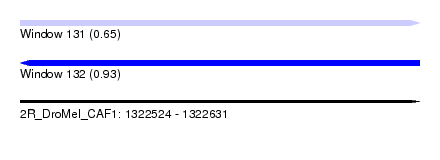
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,322,524 – 1,322,631 |
| Length | 107 |
| Max. P | 0.934196 |

| Location | 1,322,524 – 1,322,631 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
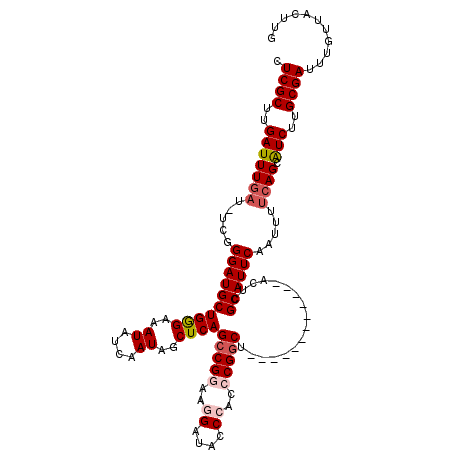
| Reading direction | forward |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -22.94 |
| Energy contribution | -24.02 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1322524 107 + 20766785 CAAGUAACAAAUCGCAAGAUGCUGAAAAUUGAAUGCGAGU------------AGCCGGGUGGGUAUCCUUCCGGCUGAGCUAUUGAUAUUUCCCAGCAUCCCGA-AUCAAAUCAAGCGAG ...........((((..(((((((.(((((.(((((...(------------(((((((.((....)).)))))))).)).))).).))))..)))))))..((-......))..)))). ( -34.80) >DroSec_CAF1 41115 107 + 1 CAAGUAACAAAUCGCAAGAUGCUGAAAAUUGAAUGCGAGU------------AGCCGGGUGGGUAUCCUUCCGGCUGAGCUAUUGAUAUUUCCCAGCAUCCCGA-AUAAAAUCAAGCGAG ...........((((..(((((((.(((((.(((((...(------------(((((((.((....)).)))))))).)).))).).))))..)))))))..((-......))..)))). ( -34.80) >DroSim_CAF1 20150 107 + 1 CAAGUAACAAAUCGCAAGAUGCUGAAAAUUGAAUGCGAGU------------AGCCGGGUGGGUAUCCUUCCGGCUGAGCUAUUGAUAUUUCCCAGCAUCCCGA-AUCAAAUCAAGCGAG ...........((((..(((((((.(((((.(((((...(------------(((((((.((....)).)))))))).)).))).).))))..)))))))..((-......))..)))). ( -34.80) >DroEre_CAF1 22446 116 + 1 CAAGUAACAAAUCGCAGGACGCUGGAAAUUGAAUGCGAGUGCGAGUGCGAGUAGUCG----GGAAUCCUUCCGGCUGAGCCAUUGAUAUUUCCCAGCAUCCCGAAAUCAAAUCAAGCGAG ...........((((.(((.(((((........(((....)))((((.(..((((((----(((....)))))))))..)))))........))))).))).((.......))..)))). ( -34.30) >DroYak_CAF1 8522 104 + 1 CAAGUAACAAAUCGCAAGAUGCUGAAAAUUGAAUGCGAGU------------GGCCG----GGAAUCCUUCCGGCUGAGCCAUUGAUAUUUCUCAGCAUCCCGAAAUCAAAUCAAGCGAU ..........(((((..(((((((((((((.((((.(..(------------(((((----(((....)))))))))..))))).).)))).))))))))..((.......))..))))) ( -35.30) >consensus CAAGUAACAAAUCGCAAGAUGCUGAAAAUUGAAUGCGAGU____________AGCCGGGUGGGUAUCCUUCCGGCUGAGCUAUUGAUAUUUCCCAGCAUCCCGA_AUCAAAUCAAGCGAG ...........((((..(((((((....(((....)))..............(((((((.((....)).))))))).................)))))))..((.......))..)))). (-22.94 = -24.02 + 1.08)

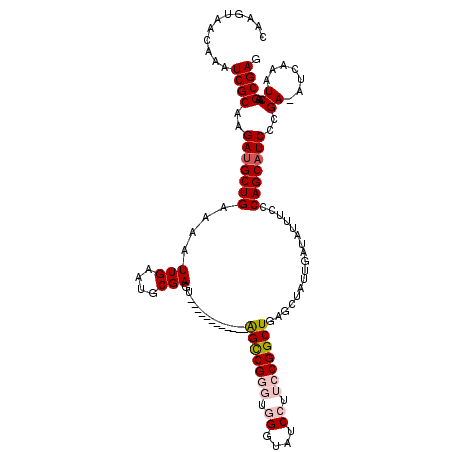
| Location | 1,322,524 – 1,322,631 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -25.96 |
| Energy contribution | -27.04 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1322524 107 - 20766785 CUCGCUUGAUUUGAU-UCGGGAUGCUGGGAAAUAUCAAUAGCUCAGCCGGAAGGAUACCCACCCGGCU------------ACUCGCAUUCAAUUUUCAGCAUCUUGCGAUUUGUUACUUG .((((..((......-))(((((((((..((.....(((.((..((((((..((....))..))))))------------....)))))...))..)))))))))))))........... ( -33.40) >DroSec_CAF1 41115 107 - 1 CUCGCUUGAUUUUAU-UCGGGAUGCUGGGAAAUAUCAAUAGCUCAGCCGGAAGGAUACCCACCCGGCU------------ACUCGCAUUCAAUUUUCAGCAUCUUGCGAUUUGUUACUUG .((((..((......-))(((((((((..((.....(((.((..((((((..((....))..))))))------------....)))))...))..)))))))))))))........... ( -33.40) >DroSim_CAF1 20150 107 - 1 CUCGCUUGAUUUGAU-UCGGGAUGCUGGGAAAUAUCAAUAGCUCAGCCGGAAGGAUACCCACCCGGCU------------ACUCGCAUUCAAUUUUCAGCAUCUUGCGAUUUGUUACUUG .((((..((......-))(((((((((..((.....(((.((..((((((..((....))..))))))------------....)))))...))..)))))))))))))........... ( -33.40) >DroEre_CAF1 22446 116 - 1 CUCGCUUGAUUUGAUUUCGGGAUGCUGGGAAAUAUCAAUGGCUCAGCCGGAAGGAUUCC----CGACUACUCGCACUCGCACUCGCAUUCAAUUUCCAGCGUCCUGCGAUUUGUUACUUG ......((((..((((.((((((((((((((.......(((.....((....))....)----)).......((....))............)))))))))))))).)))).)))).... ( -34.10) >DroYak_CAF1 8522 104 - 1 AUCGCUUGAUUUGAUUUCGGGAUGCUGAGAAAUAUCAAUGGCUCAGCCGGAAGGAUUCC----CGGCC------------ACUCGCAUUCAAUUUUCAGCAUCUUGCGAUUUGUUACUUG (((((..(((((((.....((((((((((..((....))..))))(((((........)----)))).------------....)))))).....)))).)))..))))).......... ( -32.90) >consensus CUCGCUUGAUUUGAU_UCGGGAUGCUGGGAAAUAUCAAUAGCUCAGCCGGAAGGAUACCCACCCGGCU____________ACUCGCAUUCAAUUUUCAGCAUCUUGCGAUUUGUUACUUG .((((..(((((((.....((((((((((..((....))..))))(((((..((....))..))))).................)))))).....)))).)))..))))........... (-25.96 = -27.04 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:29 2006