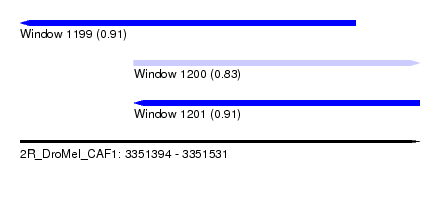
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,351,394 – 3,351,531 |
| Length | 137 |
| Max. P | 0.908909 |

| Location | 3,351,394 – 3,351,509 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.32 |
| Mean single sequence MFE | -36.31 |
| Consensus MFE | -23.98 |
| Energy contribution | -25.09 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3351394 115 - 20766785 -AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA-AAGUGCGAGUUUCAGCCACUUUUGGGAGU-UCAGCCCAGCA -............(((((.....((((((((((((((....)))))))))))).))((((((..(...........-..)..))))))))))).....(((((...-....))))).. ( -37.42) >DroVir_CAF1 11582 93 - 1 AAAAGUAUUUCACACUGACGCUUUGGUUUAACGUCA-ACAAUUGACGUUAAGCGCGAACUUGUGCGUUGUUGAUGA-AAAUGCCAACGAGAAGCA----------------------- ....((.((((.....((((((((((((((((((((-.....))))))))))).)))).....)))))((((((..-..))..)))))))).)).----------------------- ( -26.90) >DroSec_CAF1 12020 116 - 1 -AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA-AAGUGCGAGUUUCAGCCACUUUUGGUAGUUUCAGCCCAGCA -............(((((.((..((((((((((((((....)))))))))))).))((((((..(...........-..)..))))))...((((....)))).)).)))))...... ( -36.72) >DroEre_CAF1 3200 115 - 1 -AAAGUAUUUCACACUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA-AAGUGCGAGUUCCAGGCAGAUUUGGGAGU-UCAGCCCAGCA -..................((((((((((((((((((....))))))))))))...((((((..(...........-..)..))))))))))))....(((((...-....))))).. ( -40.92) >DroYak_CAF1 6627 115 - 1 -AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA-AAGUGCGAGUUCCAAGCAGUUUUGGGAGC-UCAGCCCAUCA -............(((((.((.(((((((((((((((....)))))))))))).))).((((..(...........-..)..))))(((((((....)))))))))-)))))...... ( -38.32) >DroAna_CAF1 5378 116 - 1 -AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGGGCGACGGUGAUAAAAAGUGCCAGCUUCAAGUAGACUUGACAGU-UCCUCCUUUUA -...........(((((.(((((((((((((((((((....)))))))))))).))).....)))).)))))...(((((....(((((((((....))))).)))-)....))))). ( -37.60) >consensus _AAAGUAUUUCACGCUGACGCCUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA_AAGUGCGAGUUUCAAGCAGUUUUGGGAGU_UCAGCCCAGCA ...................((((((((((((((((((....))))))))))))...(((((((((..............)))))))))))))))........................ (-23.98 = -25.09 + 1.11)


| Location | 3,351,433 – 3,351,531 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3351433 98 + 20766785 U-UUAUUAUCGUCGCACAAGUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUUACCC-----G-----AGCAG--- .-........(((((....).))))((((((((((((....))))))))............((((((((((....))-))))))))........-----)-----)))..--- ( -31.30) >DroVir_CAF1 11599 96 + 1 U-UCAUCAACAACGCACAAGUUCGCGCUUAACGUCAAUUGU-UGACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUUCAUGCCGCAUUACCC-----G-----CGG----- .-....................((((.(((((((((.....-)))))))))......(((...((((((((.....)))))))))))......)-----)-----)).----- ( -25.10) >DroPse_CAF1 8328 101 + 1 U-UUAUCAUCAACGCACAAGUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUU-UCAUGCUGUAUUACCC-----C-----AUGAAAGG .-...((((((((......))))..((((((((((((....))))))))........))))((((((((((....))-))))))))........-----.-----)))).... ( -27.00) >DroGri_CAF1 5510 101 + 1 C-UCAUCAGCAACGCACAAGUUCGCGCUUAACGUCAAUUGU-UGACGUUAAACCAAAGCGUCAGUGUGAAAUACUUUUCCAUGGCGCAUUACCU-----UGUUUCCCG----- .-.....((((((((........))).(((((((((.....-)))))))))......((((((.((.((((....))))))))))))......)-----)))).....----- ( -25.60) >DroAna_CAF1 5417 104 + 1 UUUUAUCACCGUCGCCCAAGUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCCAAGCGUCAGCGUGAAAUACUUU-UCAUGCUGUAUUACCCAGUGGG-----GGCGG--- ........(((((.((((...(((((((.((((((((....)))))))).......)))).((((((((((....))-))))))))......))).))))-----)))))--- ( -40.40) >DroPer_CAF1 9262 101 + 1 U-UUAUCAUCAACGCACAAGUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGAGUCGGCGUGAAAUACUUU-UCAUGCUGUUUUACCC-----C-----AUGAAAGG .-...((((((((......))))..((((((((((((....))))))))........))))((((((((((....))-))))))))........-----.-----)))).... ( -27.00) >consensus U_UUAUCAUCAACGCACAAGUUGGCGCUCAACGUCAAUUGUUUGACGUUAAACCAAAGCGUCAGCGUGAAAUACUUU_UCAUGCUGUAUUACCC_____G_____AGGA____ ............(((........)))...((((((((....))))))))............((((((((.(.....).))))))))........................... (-16.40 = -17.07 + 0.67)

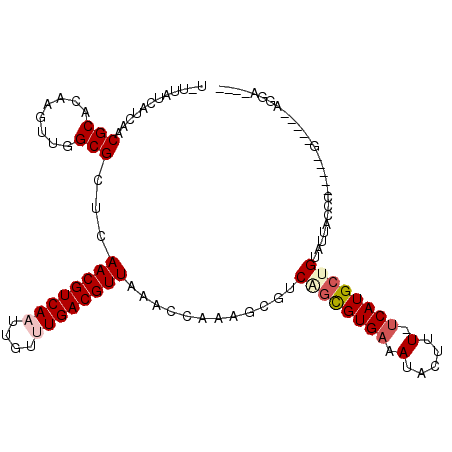
| Location | 3,351,433 – 3,351,531 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -32.17 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3351433 98 - 20766785 ---CUGCU-----C-----GGGUAAUACAGCAUGA-AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGACGAUAAUAA-A ---.(((.-----(-----((((....((((.(((-((....))))).)))).....(((((((((((((((....)))))))))))).)))))))).)))..........-. ( -35.30) >DroVir_CAF1 11599 96 - 1 -----CCG-----C-----GGGUAAUGCGGCAUGAAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUCA-ACAAUUGACGUUAAGCGCGAACUUGUGCGUUGUUGAUGA-A -----(((-----(-----(.....))))).............((((((..((((((((((((((((((((-.....))))))))))).)))).....)))))..)).)))-) ( -30.00) >DroPse_CAF1 8328 101 - 1 CCUUUCAU-----G-----GGGUAAUACAGCAUGA-AAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGUUGAUGAUAA-A ..((((((-----(-----..(.....)..)))))-))..(((((((((((((....(((((((((((((((....))))))))))..))))).))).))).))).)))).-. ( -28.50) >DroGri_CAF1 5510 101 - 1 -----CGGGAAACA-----AGGUAAUGCGCCAUGGAAAAGUAUUUCACACUGACGCUUUGGUUUAACGUCA-ACAAUUGACGUUAAGCGCGAACUUGUGCGUUGCUGAUGA-G -----..(....).-----.((((((((((((((((((....)))).)).)).(((....(((((((((((-.....)))))))))))))).....)))))))))).....-. ( -32.10) >DroAna_CAF1 5417 104 - 1 ---CCGCC-----CCCACUGGGUAAUACAGCAUGA-AAAGUAUUUCACGCUGACGCUUGGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGGGCGACGGUGAUAAAA ---.((((-----(...(..(((....((((.(((-((....))))).))))..)))..)((((((((((((....))))))))))))........)))))............ ( -38.60) >DroPer_CAF1 9262 101 - 1 CCUUUCAU-----G-----GGGUAAAACAGCAUGA-AAAGUAUUUCACGCCGACUCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGUUGAUGAUAA-A ..((((((-----(-----..(.....)..)))))-))..(((((((((((((....(((((((((((((((....))))))))))..))))).))).))).))).)))).-. ( -28.50) >consensus ____UCCU_____C_____GGGUAAUACAGCAUGA_AAAGUAUUUCACGCUGACGCUUUGGUUUAACGUCAAACAAUUGACGUUGAGCGCCAACUUGUGCGUUGAUGAUAA_A ....................((.(((((...........))))))).((.((((((((((((((((((((((....)))))))))))).)))).....)))))).))...... (-20.18 = -20.52 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:29 2006