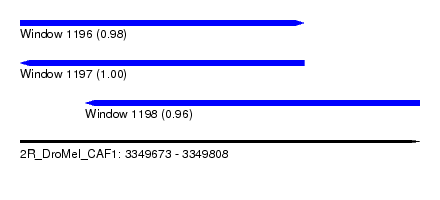
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,349,673 – 3,349,808 |
| Length | 135 |
| Max. P | 0.999979 |

| Location | 3,349,673 – 3,349,769 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3349673 96 + 20766785 UUUGCCAUUCAGACUUUCCCAGAAAUCUCAAUUAGAUUGACACUCCUGCCAUGGCUGCCAUGGCAAACCACUUGGAUUGGA-----UCCGAUUGGCCCAAC ...((((.((.((((.(((.((.(((((.....)))))........((((((((...)))))))).....)).)))..)).-----)).)).))))..... ( -27.80) >DroSec_CAF1 10224 96 + 1 UUUGCCAUUCAGACUUUCCCAGAAAUCUCAAUUAGAUUGACACUCCUGCCAUGGCUGCCAUGGCAAACCACUUGGAUUGGA-----UCCGAUUGGCCCAAC ...((((.((.((((.(((.((.(((((.....)))))........((((((((...)))))))).....)).)))..)).-----)).)).))))..... ( -27.80) >DroSim_CAF1 3369 96 + 1 UUUGCCAUUCAGACUUUCCCAGAAAUCUCAAUUAGAUUGACACUCCUGCCAUGGCUGCCAUGGCAAACCACUUGGAUUGGA-----UCCGAUUGGUCCAAC ...((((.((.((((.(((.((.(((((.....)))))........((((((((...)))))))).....)).)))..)).-----)).)).))))..... ( -25.90) >DroEre_CAF1 1474 101 + 1 UUUGCCAUUCAGACUUUCCCAGAAAUCUCAAUUAGAUUGACACUCCUACCAUGGCUGCCAUGGCAAUGCAAUUGGAUUGGAUUUGUUCGGAUUGGCCCGAC ...((((.((.((.......(.(((((.(((((.(((((.((......((((((...))))))...))))))).)))))))))).))).)).))))..... ( -24.81) >DroYak_CAF1 4854 96 + 1 UUUGCCAUUCAGACUUGCCCAGAAAUCUCAAUUAGAUUGACAGUCCUGCCACGGCUGCCAUGGCAAUGCACUUGGAUUGAA-----UCGGAUUGGCUGAAC ...((((.((.((..........(((((.....)))))..(((((((((((.((...)).)))))........))))))..-----)).)).))))..... ( -23.90) >consensus UUUGCCAUUCAGACUUUCCCAGAAAUCUCAAUUAGAUUGACACUCCUGCCAUGGCUGCCAUGGCAAACCACUUGGAUUGGA_____UCCGAUUGGCCCAAC ...((((.((.((.....((((.(((((.....))))).....(((((((((((...))))))))........)))))))......)).)).))))..... (-22.06 = -22.50 + 0.44)

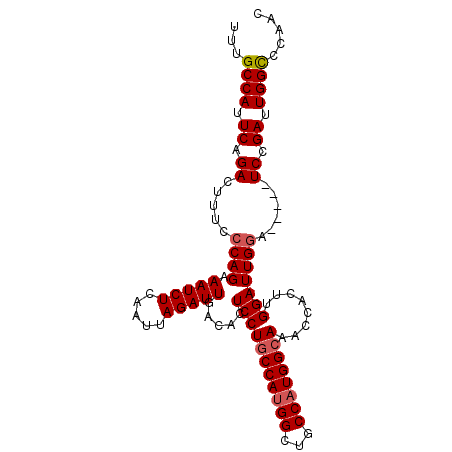
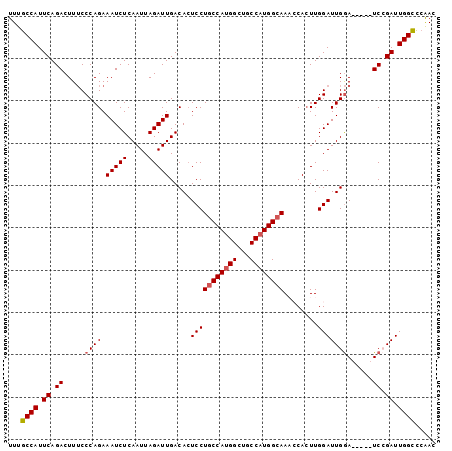
| Location | 3,349,673 – 3,349,769 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3349673 96 - 20766785 GUUGGGCCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUUCUGGGAAAGUCUGAAUGGCAAA .....((((.(((((-----(....(((..((((((.((....))))))))..(((((((.(((((...))))).))))))))))..)))))).))))... ( -37.50) >DroSec_CAF1 10224 96 - 1 GUUGGGCCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUUCUGGGAAAGUCUGAAUGGCAAA .....((((.(((((-----(....(((..((((((.((....))))))))..(((((((.(((((...))))).))))))))))..)))))).))))... ( -37.50) >DroSim_CAF1 3369 96 - 1 GUUGGACCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUUCUGGGAAAGUCUGAAUGGCAAA ....(.(((.(((((-----(....(((..((((((.((....))))))))..(((((((.(((((...))))).))))))))))..)))))).))))... ( -33.70) >DroEre_CAF1 1474 101 - 1 GUCGGGCCAAUCCGAACAAAUCCAAUCCAAUUGCAUUGCCAUGGCAGCCAUGGUAGGAGUGUCAAUCUAAUUGAGAUUUCUGGGAAAGUCUGAAUGGCAAA .....((((.((.((.....((((............((((((((...))))))))(((((.(((((...))))).)))))))))....)).)).))))... ( -29.80) >DroYak_CAF1 4854 96 - 1 GUUCAGCCAAUCCGA-----UUCAAUCCAAGUGCAUUGCCAUGGCAGCCGUGGCAGGACUGUCAAUCUAAUUGAGAUUUCUGGGCAAGUCUGAAUGGCAAA .....((((.((.((-----((...(((((((.(..((((((((...))))))))).)))...(((((.....)))))..))))..)))).)).))))... ( -29.60) >consensus GUUGGGCCAAUCGGA_____UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUUCUGGGAAAGUCUGAAUGGCAAA .....((((.(((((.....((((............((((((((...))))))))(((((.(((((...))))).)))))))))....))))).))))... (-31.70 = -32.02 + 0.32)

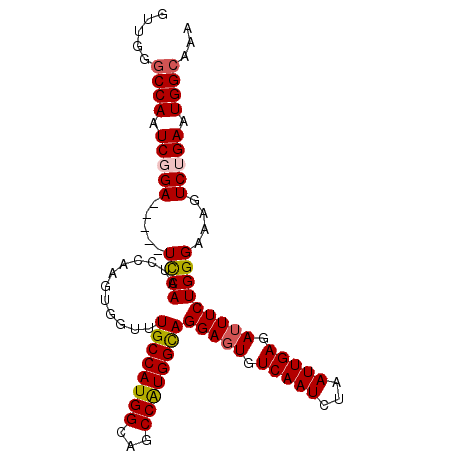
| Location | 3,349,695 – 3,349,808 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.957213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
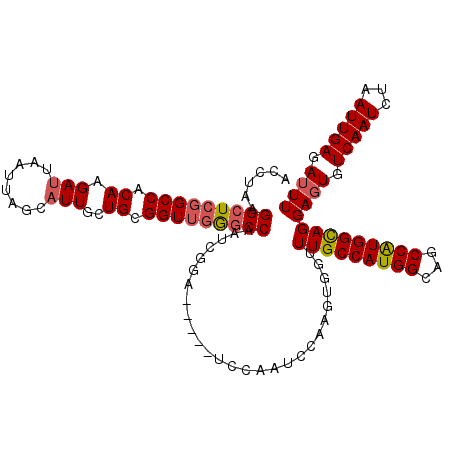
>2R_DroMel_CAF1 3349695 113 - 20766785 ACCUAAGGCUCGGCCACAAGAUUAAUUAGCAUUGCUGCGGUUGGGCCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUU (((...(((((((((.((.(((........)))..)).)))))))))....(((-----(...))))....)))(((((((((...)))))))))((((.(((((...))))).)))) ( -38.20) >DroSec_CAF1 10246 113 - 1 ACCUAAGGCUCGGCCACAAGAUUAAUUAGCAUUGCUGCGGUUGGGCCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUU (((...(((((((((.((.(((........)))..)).)))))))))....(((-----(...))))....)))(((((((((...)))))))))((((.(((((...))))).)))) ( -38.20) >DroSim_CAF1 3391 113 - 1 ACCUAAGGCUCGGCCACAAGAUUAAUGAGCAUUGCUGCGGUUGGACCAAUCGGA-----UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUU .......((((.(((((...........(((....)))(((((((((....)).-----)))))))...)))))(((((((((...)))))))))))))...(((((.....))))). ( -38.70) >DroEre_CAF1 1496 118 - 1 AUCUAAGGCUCCGCCACAAGAUUAAUUAGCAUUGCUGUGGUCGGGCCAAUCCGAACAAAUCCAAUCCAAUUGCAUUGCCAUGGCAGCCAUGGUAGGAGUGUCAAUCUAAUUGAGAUUU ......(((((.((((((.(((........)))..)))))).))))).........(((((((((...(((((((((((((((...))))))))...))).))))...)))).))))) ( -36.10) >DroYak_CAF1 4876 113 - 1 GUCCAAGGCUCGGCCACAAGAUUAAUUAGCUUUGCUGCGGUUCAGCCAAUCCGA-----UUCAAUCCAAGUGCAUUGCCAUGGCAGCCGUGGCAGGACUGUCAAUCUAAUUGAGAUUU (((((((((...)))...(((((...(((..((((..(((((..((((...(((-----(.((.......)).))))...)))))))))..))))..)))..)))))..))).))).. ( -32.80) >consensus ACCUAAGGCUCGGCCACAAGAUUAAUUAGCAUUGCUGCGGUUGGGCCAAUCGGA_____UCCAAUCCAAGUGGUUUGCCAUGGCAGCCAUGGCAGGAGUGUCAAUCUAAUUGAGAUUU ......(((((((((.((.(((........)))..)).)))))))))...........................(((((((((...)))))))))((((.(((((...))))).)))) (-30.08 = -30.60 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:26 2006