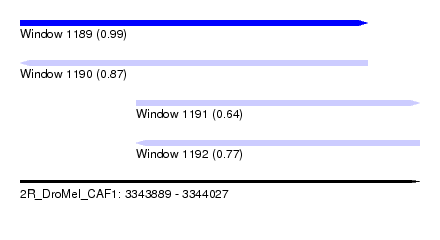
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,343,889 – 3,344,027 |
| Length | 138 |
| Max. P | 0.993711 |

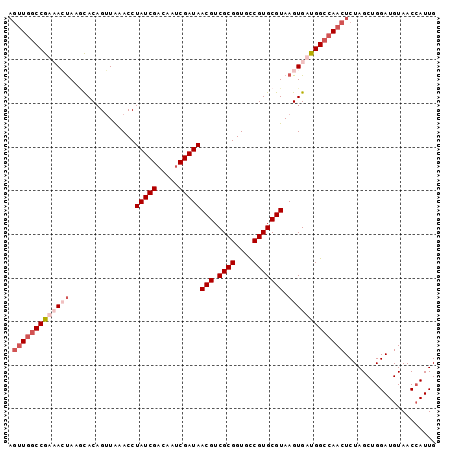
| Location | 3,343,889 – 3,344,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -26.27 |
| Energy contribution | -28.90 |
| Covariance contribution | 2.63 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
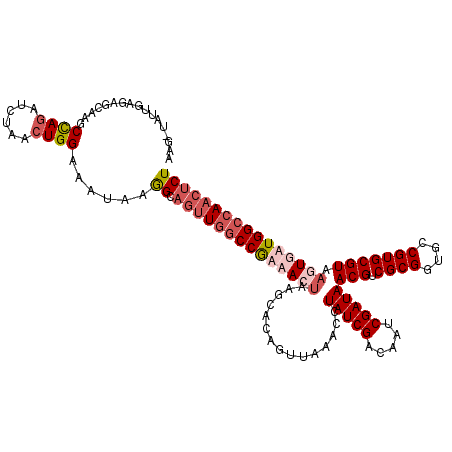
>2R_DroMel_CAF1 3343889 120 + 20766785 AAGCUAUUGAGCGCAAGCUAAAUCUAACUGGAAUUAAGGCAGUUGGCCGUAACUAAGCACAGGUGAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCU ..(((....(((....)))...(((....))).....)))((((((((((.(((..((((..((((...(((((.....)))))...))))(....)))))...))).)))))))))).. ( -43.50) >DroSim_CAF1 15502 120 + 1 AAGCUAUUUAGCACAAGCUAGAUCUAACUGGAAUUAAGGCAGUUGGCCGUAACUAAUCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCU ..(((.((((((....))))))(((....))).....)))((((((((((.(((...............(((((.....)))))(((.((((....))))))).))).)))))))))).. ( -39.20) >DroEre_CAF1 15616 112 + 1 --------GAAAGCAAGCCAGAUCUAUCUGGAAAUAAAGCAGUUGGCCGAUAGUACGCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGUCGUGCGUAAGUGAUGGCCAACUCU --------....((...(((((....))))).......))(((((((((.((.(((((((((......))...((((..((((...))))...)))))))))))..)).))))))))).. ( -38.10) >DroYak_CAF1 16241 117 + 1 AGG-ACCAAAGAGCAAGCCAGAUUUAACUGGAAAUAAGGCCAAU-ACCAAUAGCA-AUACAGUUAAACCUAUCGUCAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCU ...-.....((((....((((......))))......(((((.(-((...((((.-.....))))....(((((.....)))))(((.((((....)))))))..))).)))))..)))) ( -31.60) >consensus AAG_UAUUGAGAGCAAGCCAGAUCUAACUGGAAAUAAGGCAGUUGGCCGAAACUAAGCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCU .................((((......))))......((.((((((((((((((...............(((((.....)))))(((.((((....))))))).)))))))))))))))) (-26.27 = -28.90 + 2.63)

| Location | 3,343,889 – 3,344,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -22.85 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3343889 120 - 20766785 AGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUCACCUGUGCUUAGUUACGGCCAACUGCCUUAAUUCCAGUUAGAUUUAGCUUGCGCUCAAUAGCUU ..((((((((...(((...((((((.(((.(((((........)))))...)))....))))))..)))...))))))))...........(((((.....(((....)))...))))). ( -37.40) >DroSim_CAF1 15502 120 - 1 AGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGAUUAGUUACGGCCAACUGCCUUAAUUCCAGUUAGAUCUAGCUUGUGCUAAAUAGCUU ..((((((((...(((....(((((.(((.(((((........)))))...)))....)))))...)))...))))))))...........(((((....((((....))))..))))). ( -36.90) >DroEre_CAF1 15616 112 - 1 AGAGUUGGCCAUCACUUACGCACGACACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGCGUACUAUCGGCCAACUGCUUUAUUUCCAGAUAGAUCUGGCUUGCUUUC-------- ..((((((((......((((((((..(((.(((((........)))))...))).....)))))))).....))))))))((.......(((((....)))))...))....-------- ( -38.30) >DroYak_CAF1 16241 117 - 1 AGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGACGAUAGGUUUAACUGUAU-UGCUAUUGGU-AUUGGCCUUAUUUCCAGUUAAAUCUGGCUUGCUCUUUGGU-CCU (((((.((((........(((........))).(((((.....)))))..(((((((((((((.-.((((.....-..))))..))....))))))))))))))).))))).....-... ( -33.50) >consensus AGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGCUUACUAACGGCCAACUGCCUUAAUUCCAGUUAGAUCUAGCUUGCGCUCAAUA_CUU ..((((((((...(((...((((((.(((.(((((........)))))...)))....))))))..)))...)))))))).........((((......))))................. (-22.85 = -24.73 + 1.87)

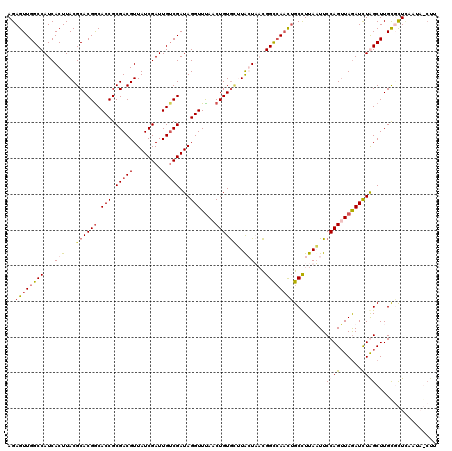
| Location | 3,343,929 – 3,344,027 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -22.99 |
| Energy contribution | -25.55 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
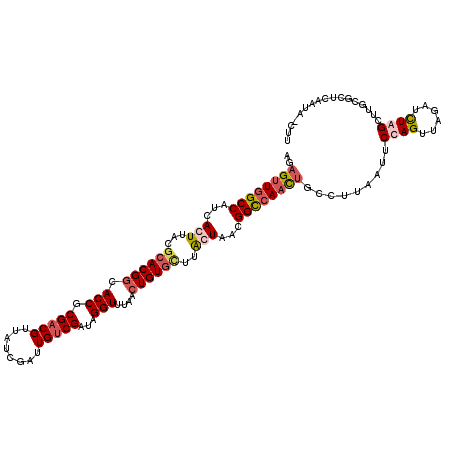
>2R_DroMel_CAF1 3343929 98 + 20766785 AGUUGGCCGUAACUAAGCACAGGUGAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAGCUGGAUGUAACCAUUG ((((((((((.(((..((((..((((...(((((.....)))))...))))(....)))))...))).)))))))))).....(((......)))... ( -35.50) >DroSim_CAF1 15542 98 + 1 AGUUGGCCGUAACUAAUCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAGCUGGAUGUAACCAUUG ((((((((((.(((...............(((((.....)))))(((.((((....))))))).))).)))))))))).....(((......)))... ( -31.90) >DroEre_CAF1 15648 98 + 1 AGUUGGCCGAUAGUACGCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGUCGUGCGUAAGUGAUGGCCAACUCUAGCUGGAUGUAACGAUUG (((((((((.((.(((((((((......))...((((..((((...))))...)))))))))))..)).))))))))).................... ( -31.30) >DroYak_CAF1 16280 96 + 1 CAAU-ACCAAUAGCA-AUACAGUUAAACCUAUCGUCAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAGCUGGAUGUAACCAUUG ....-.....((((.-.....))))....(((((.....)))))(((((.(((((((((((....)).)))))........)))))))))........ ( -20.60) >consensus AGUUGGCCGAAACUAAGCACAGUUAAACCUAUCGACAAUCGAUAACGUCGCGGUGCCGUGCGUAAGUGAUGGCCAACUCUAGCUGGAUGUAACCAUUG .(((((((((((((...............(((((.....)))))(((.((((....))))))).)))))))))))))..................... (-22.99 = -25.55 + 2.56)

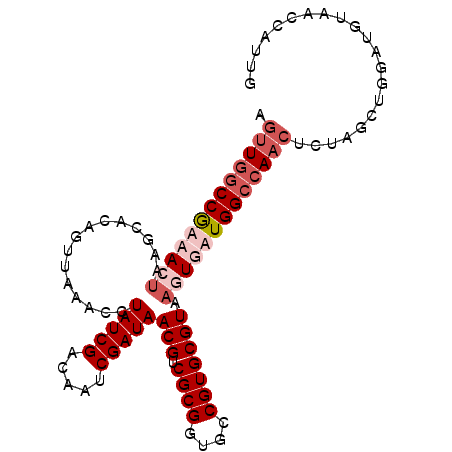
| Location | 3,343,929 – 3,344,027 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.63 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -22.29 |
| Energy contribution | -23.85 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3343929 98 - 20766785 CAAUGGUUACAUCCAGCUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUCACCUGUGCUUAGUUACGGCCAACU ...(((......))).....((((((((...(((...((((((.(((.(((((........)))))...)))....))))))..)))...)))))))) ( -34.70) >DroSim_CAF1 15542 98 - 1 CAAUGGUUACAUCCAGCUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGAUUAGUUACGGCCAACU ...(((......))).....((((((((...(((....(((((.(((.(((((........)))))...)))....)))))...)))...)))))))) ( -32.00) >DroEre_CAF1 15648 98 - 1 CAAUCGUUACAUCCAGCUAGAGUUGGCCAUCACUUACGCACGACACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGCGUACUAUCGGCCAACU ....................((((((((......((((((((..(((.(((((........)))))...))).....)))))))).....)))))))) ( -32.20) >DroYak_CAF1 16280 96 - 1 CAAUGGUUACAUCCAGCUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGACGAUAGGUUUAACUGUAU-UGCUAUUGGU-AUUG (((((((((((.(((((....))))).....((((((((........))).(((((.....))))).))))).....)))).-.)))))))..-.... ( -22.60) >consensus CAAUGGUUACAUCCAGCUAGAGUUGGCCAUCACUUACGCACGGCACCGCGACGUUAUCGAUUGUCGAUAGGUUUAACUGUGCUUACUAACGGCCAACU ...(((((......)))))..(((((((...(((...((((((.(((.(((((........)))))...)))....))))))..)))...))))))). (-22.29 = -23.85 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:20 2006