
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,321,061 – 3,321,177 |
| Length | 116 |
| Max. P | 0.614269 |

| Location | 3,321,061 – 3,321,177 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3321061 116 + 20766785 AUUGUUGUUCUCAUU----UUUUAGUUAAUGACUGGCAUGUCCAAGCUCAUGAAGGAGUGCUGGCACCAAAACCCCGACGUGCGCUUGCCAGCGCUGCGCAUCAAAAAGACUAUACAUAA .(((.(((.((((((----........)))))((..((((........))))..))((((((((((.....((......)).....))))))))))).))).)))............... ( -27.20) >DroVir_CAF1 2007 114 + 1 AUUAUAUACCU--GU----UUGUAGCUACUGGCCGGCAUGUCCAAGCUAAUGAAGGAGUGCUGGCAUCAGAAUCCGAAUGUGCGUUUGCCAGCGUUGCGCAUUAAAAAGACUAUACAUAA ..........(--((----.(((((((.(((((((((((.(((...(....)..)))))))))))..)))......((((((((..((....)).))))))))....)).))))).))). ( -31.20) >DroGri_CAF1 2232 115 + 1 CCUUUCUUUCUU-UU----AAACAGCUCCUUGCCGGCAUGUCUAAGCUAAUGAGGGAGUGCUGGCAUCAGAAUCCGAACGUUCGUUUGCCAGCGCUGCGCAUUAAAAAGACUAUACAUAA ........((((-((----.((..((((((((..(((........)))..))))))((((((((((...((((......))))...))))))))))))...)).)))))).......... ( -33.40) >DroMoj_CAF1 1982 115 + 1 AUUAUGUUUCUC-AU----CUACAGCUUCUGGCUGGCAUGUCCAAGCUAAUGAAGGAAUGCUGGCAUCAGAAUCCGAAUGUUCGCCUGCCAGCGUUGCGCAUUAAAAAGACUAUACAUAA .((((((.....-..----...((((.....))))....(((.....(((((..(.((((((((((...((((......))))...)))))))))).).)))))....)))...)))))) ( -32.90) >DroAna_CAF1 2829 115 + 1 AU--CU---CUCCUUUCUUGAUCAGCUACUGGCUGGGAUGUCUAAACUAAUGAAAGAGUGCUGGCACCAGAACCCCAACGUGCGCUUGCCAGCGCUGCGCAUCAAAAAGACCAUACAUAA ..--..---......((((..(((((.....)))))(((((.....((......))((((((((((...(.((......)).)...))))))))))..)))))...)))).......... ( -29.30) >DroPer_CAF1 1936 113 + 1 AUUGGU---UUUAUU----UUACAGCUGCUGGCCGGCAUGUCCAAGCUAAUGAAGGAGUGUUGGCACCAGAACCCAAAUGUUCGUCUGCCAGCCCUUCGCAUCAAAAAGACUAUACAUAA ..((((---(((...----....((((...(((......)))..))))...(((((...(((((((...((((......))))...))))))))))))........)))))))....... ( -30.40) >consensus AUUAUUUUUCUC_UU____UUACAGCUACUGGCCGGCAUGUCCAAGCUAAUGAAGGAGUGCUGGCACCAGAACCCGAACGUGCGCUUGCCAGCGCUGCGCAUCAAAAAGACUAUACAUAA .......................((((...(((......)))..))))..(((.(.((((((((((...(.((......)).)...)))))))))).)...)))................ (-20.15 = -19.73 + -0.41)

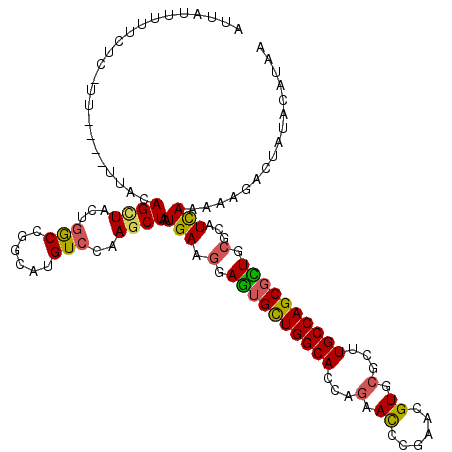
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:15 2006